
Lukas Valihrach
@lukasvalihrach
Group Leader | 🧬Single-Cell & Spatial Transcriptomics | Epigenomics | 🧠Neurobiology | Glial Cells in Health & Disease | @labgenex @IBT_CAS 🇪🇺 | ❤️ My Twins
ID: 1199388618613280768
https://www.labgenexp.eu/ 26-11-2019 18:05:15
1,1K Tweet
3,3K Followers
771 Following


Celcomen: our new tool for spatial causal disentanglement and identifiability in spatial transcriptomics to perform spatial counterfactuals such as localised gene knoc-outs! by Stathis Megas and team Wellcome Sanger Institute Cambridge Stem Cell Institute

Our recent work (led by Lauren Patel comentored by Chris Benner), is out .Nature Biotechnology rdcu.be/dTTWs. We aim to increase rigor in scientific studies. Here we show that spike-in normalization is commonly used improperly, and conclusions may actually be incorrect (1/2)





🧵 1/8 🎉 Our latest collaborative publication, "Challenges and recommendations for the translation of biomarkers of aging", has just been released in Nature Aging (Nature Aging). Biomarkers of aging predict biological age and its response to interventions but face challenges in


Our next Center for Integrated Cellular Analysis Virtual Seminar (12PM on 10/1) AbdulAbdul introduces IRISeq, a new technology to perform spatial transcriptomics in a highly scalable and cost-effective way (<$50/tissue section)- no optics or imaging required! Free registration: nygenome.zoom.us/meeting/regist…


Finally out in Nature Protocols : our group's workflows for end to end computational pathology. Compatible with UNI and other foundation models. Led by Omar S.M. El Nahhas from katherlab journal link: nature.com/articles/s4159… full text: rdcu.be/dT35l


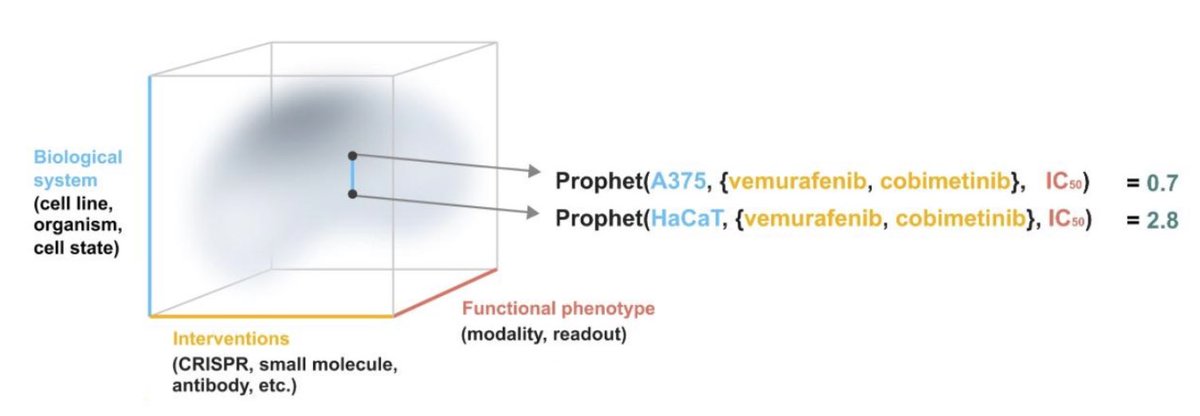
Thrilled to share our „PRedictor Of PHEnoTypes“ model Prophet! Led by Alejandro Tejada Lapuerta & Yuge Ji, Prophet is a transformer-based model that predicts outcomes for unseen experiments. It aims to understands biology by learning across assays and phenotypes over 4.7M+ experiments.


Excited to share our research on extracellular vesicle-mediated cellular crosstalk in the developing brain (organoids)!!! Great work led by the wonderful scientists Andrea Forero & Fabrizia Pipicelli cell.com/cell-reports/f…

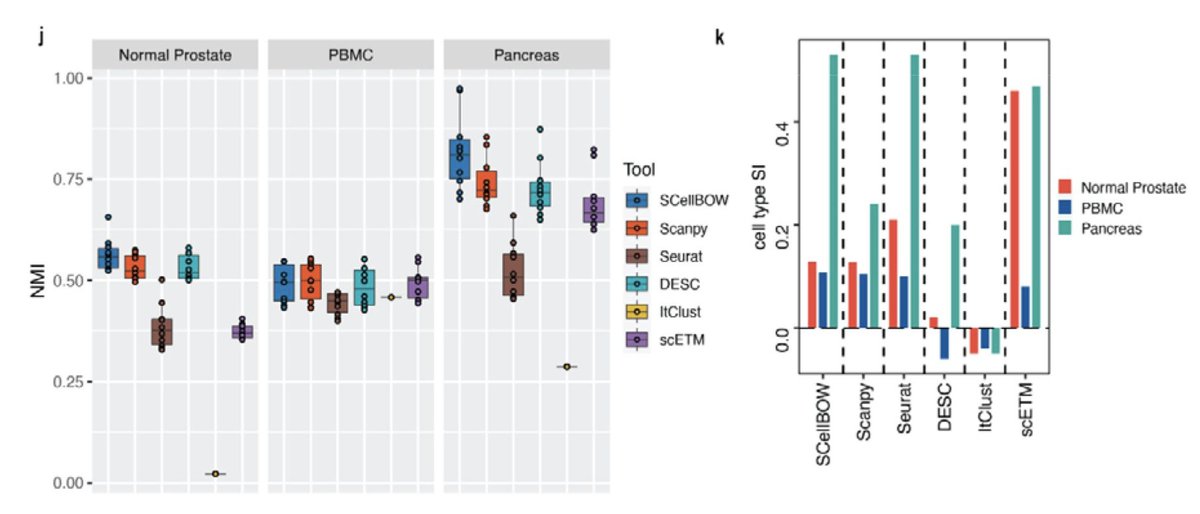
In Joseph Rich et al. biorxiv.org/content/10.110…, we assessed the Seurat and Scanpy single-cell packages, finding that they produce very different results on standard analyses. A new paper in eLife - the journal looking at clustering finds the same elifesciences.org/reviewed-prepr…

Excited to share our first preprint on a novel spatial multi-omics technology that co-profiles five modalities within the same tissue section. Great collab w/ Marek Bartosovic and Mingyao Li . Kudos to my postdoc Pengfei Guo and PhD student Liran Mao ! biorxiv.org/content/10.110…



📢Out now! Gennady Gorin, Lior Pachter and colleagues from Caltech propose a method to cluster cells from multimodal single-cell data under a self-consistent, biophysical model. nature.com/articles/s4358…


