
J.T. Neal
@jamestneal
PI @ neallab.org. Scientist building tools to understand genetic variation in health and disease. Trail runner & World’s Okayest Dad.
ID: 1482923604
http://neallab.org 04-06-2013 18:32:00
481 Tweet
782 Followers
758 Following

We’re excited to share our work developing a new gene editing approach that we’re calling #clickediting 🖱️✴️🧬. Click editors (CEs) utilize DNA-dependent polymerases to write bespoke edits into the genome. biorxiv.org/content/10.110… CGM at MGH MassGeneral News Mass General Research Institute Harvard Medical School



Of all the stories I've read or heard the past few days, Amir Tibon אמיר תיבון's is the most remarkable—capturing the horror, the failures, the heroics, the hope, and the despair of this moment in Israel. theatlantic.com/ideas/archive/…










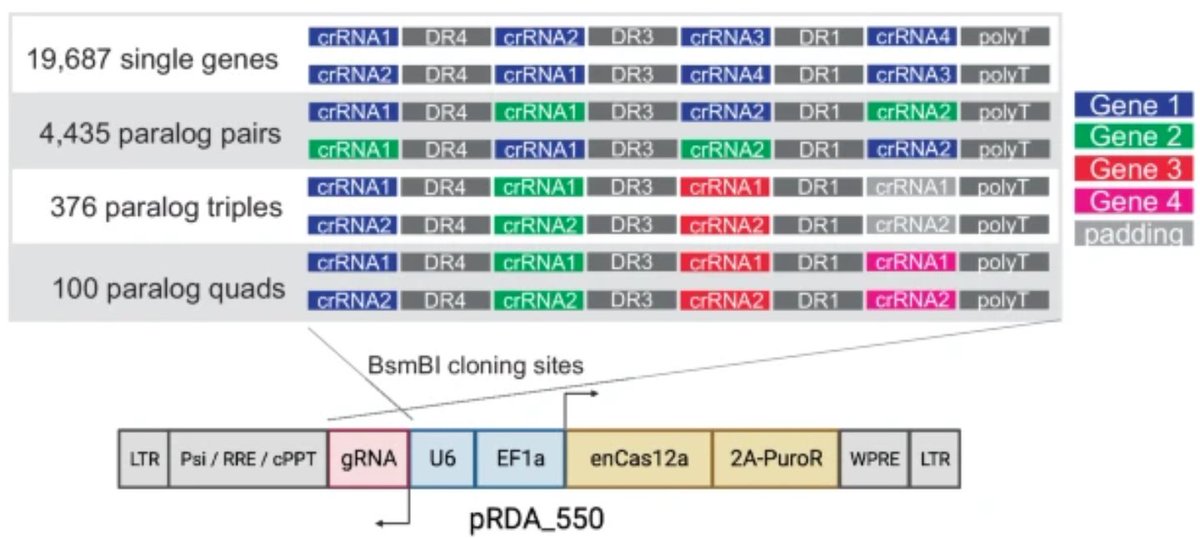
At long last, the in4mer paper is published! In4mer is our CRISPR/Cas12a platform, developed in collaboration with John Doench, where we show that expressing 4mer arrays of Cas12a guides turns out to be a highly efficient way to knock out genes singly or in combination.

ONLY 12 DAYS AWAY: The 2024 Mutational Scanning Symposium, hosted by Broad Institute , opens on May 22. If you have not already registered, please do so ASAP. Atlas of Variant Effects Alliance Harvard University Massachusetts Institute of Technology (MIT) broadinstitute.swoogo.com/mss-2024/51014…





