Robin H. van der Weide
@robinweide
Postdoc Integrative Single-Cell Functional Genomics in the @kind_jop lab (@_Hubrecht). PhD from @NKI_nl
ID: 337318314
17-07-2011 20:49:10
429 Tweet
309 Followers
263 Following

The Kind lab Hubrecht Institute describe MAbID, a multiplexing approach to uncover the genomic distributions of various epigenetic markers. They demonstrate joint measurements of six epitopes in mouse bone marrow at single-cell resolution. @kind_jop Robin H. van der Weide nature.com/articles/s4159…
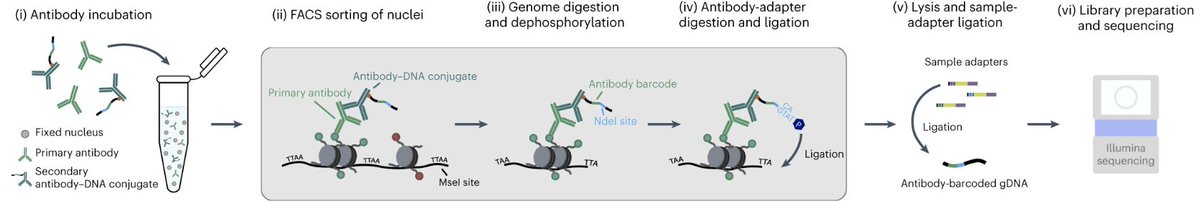

Exciting work by @kind_jop lab published in Nature Methods! Read this news item to learn more about MAbID: ru.nl/onderzoek/onde… ru.nl/en/research/re…



📢We are very excited to announce the second edition of the Dutch Embryo Model Meeting #DEMM24 Are you working with embryo models such as 2D/3D #gastruloids or #blastoids? Join our meeting and submit an abstract! oncodeinstitute.nl/events/dutch-e… Oncode Institute



Congratulations to Puck Knipscheer and @kind_jop, who have been awarded NWO Funding Vici grants for their research on resolving dangerous DNA structures and the role of chromosome organization in determining cell identity respectively.


(1/14) Excited to share our new work on ZNF143/ZFP143 by Domenic Narducci ZNF143 has been widely reported to be a general loop factor Instead we and deWitLab in a companion preprint show that ZNF143 is a TF with no loop function: biorxiv.org/content/10.110… biorxiv.org/content/10.110…
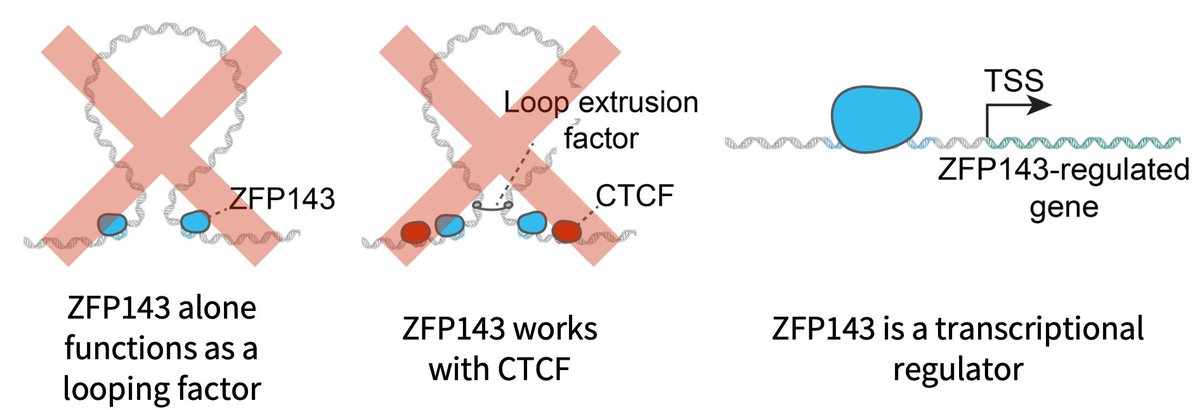

You may know ZNF143 as a looping factor. We show together with the ASHansen Lab that this is not the case! If you want to know why and what it does instead do read our thread below: biorxiv.org/content/10.110…


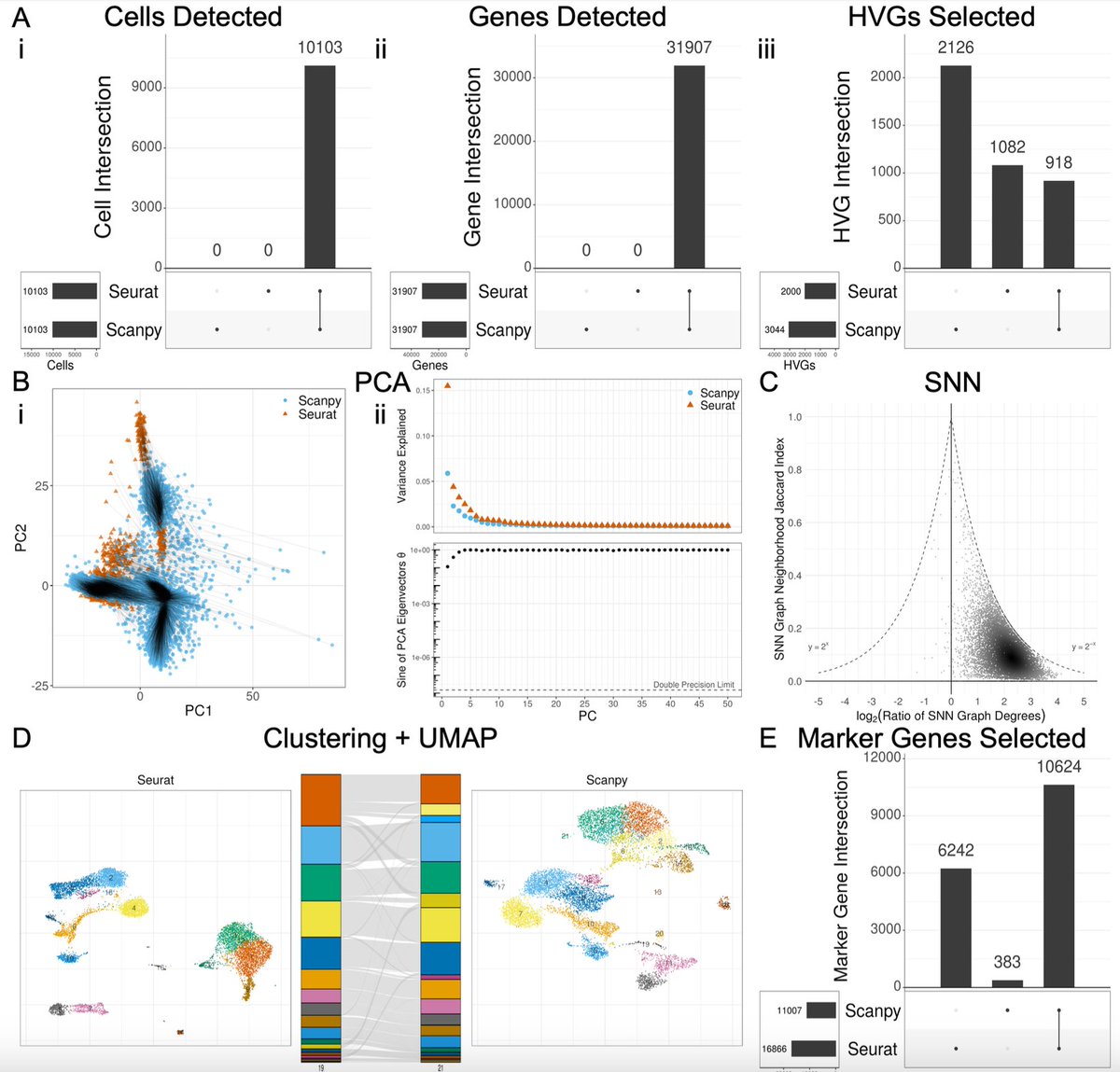
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In biorxiv.org/content/10.110… w/ Joseph Rich et al. we take a close look. The results are 👀 1/🧵

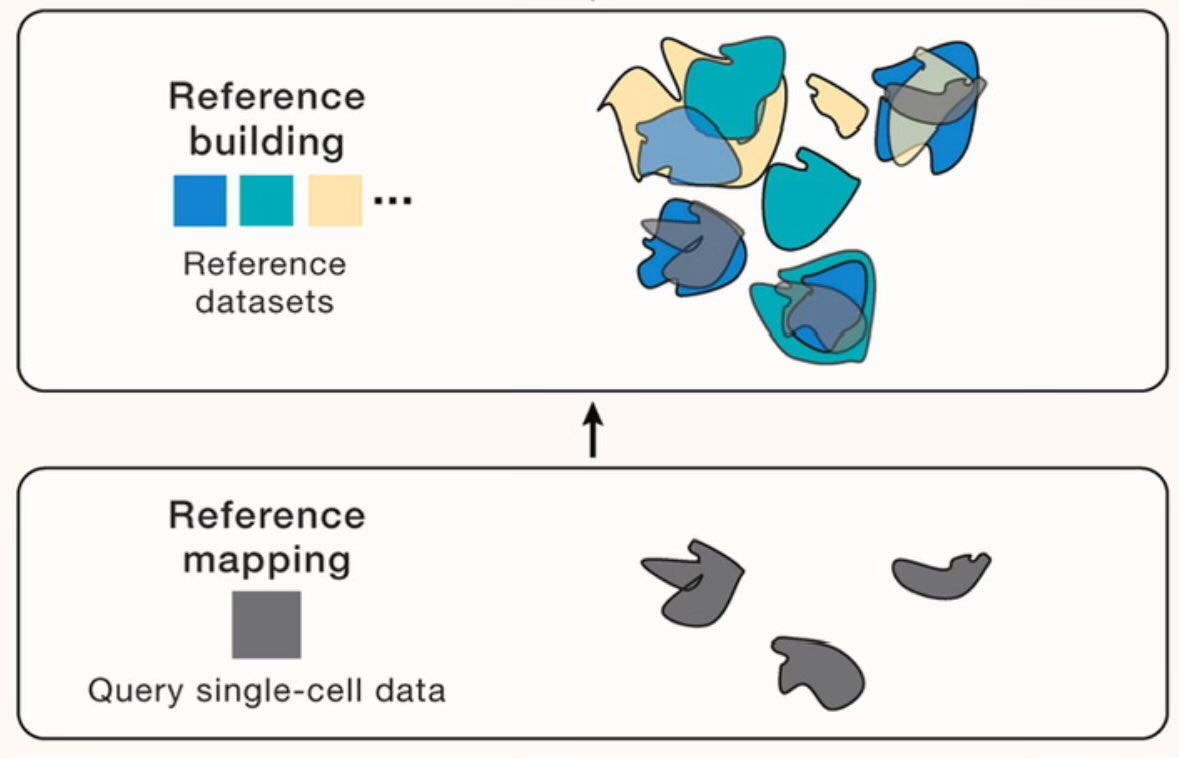
In need of an educational weekend read? Check out our new perspective Cell with Rahul Rahul Satija. Led by Mo Lotfollahi & YUHAN HAO, we outline how to automate single-cell data analysis via reference mapping. cell.com/cell/fulltext/…

How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by Grace Bower where we uncover a unique sequence signature globally associated with long-range enhancer-promoter interactions in developing limb buds: biorxiv.org/content/10.110… 1/

Out today in Nature Materials, the updated version of our work on nuclear jamming in organogenesis 👁️🧠 Thanks to the reviewers (yes, thanks 😊), we have added a lot of new data. Check below for the new results... 🧵 ➡️rdcu.be/dQDm6



Great to see Aleksander Szczurek's paper out today! A wonderful team effort with @emmy_dimitrova , Jess Kelley , and Neil Blackledge. Have a look if you are interested in how #Polycomb controls transcription to repress gene expression. rdcu.be/dTD3t

So thrilled to see our work on regulation of transcriptional repression in lamina-associated published in The EMBO Journal. Thanks to all the authors!! Thanks to all the authors for their fantastic contribution. embopress.org/doi/full/10.10…