Nicholas Ho
@eigennick
PhD Student @CMUCompBio @SCSatCMU | Machine Learning and AI4Bio Researcher | Yoyo enthusiast 🪀
ID: 1407875772078039040
http://nickdst.github.io 24-06-2021 01:38:45
52 Tweet
117 Takipçi
503 Takip Edilen

My amazing PhD student Wendy Yang Muyu "Wendy" Yang is graduating this summer & seeking industry R&D roles! She's published in ISMB, Nature Methods, and interned at Genentech. Strong in #AI/#ML for gene regulation. Looking for top AI+bio talent? Contact Wendy: [email protected]














📡 Join us for the next #FM4Bio Seminar on Sept 17 at 9 AM PT featuring Haotian Cui Haotian will present “Large Models for Single-Cell Omics and Drug Discovery: Data, Pretraining, and Closed-Loop Environment.” Save your spot → genbio.ai/fm4bio-seminar/


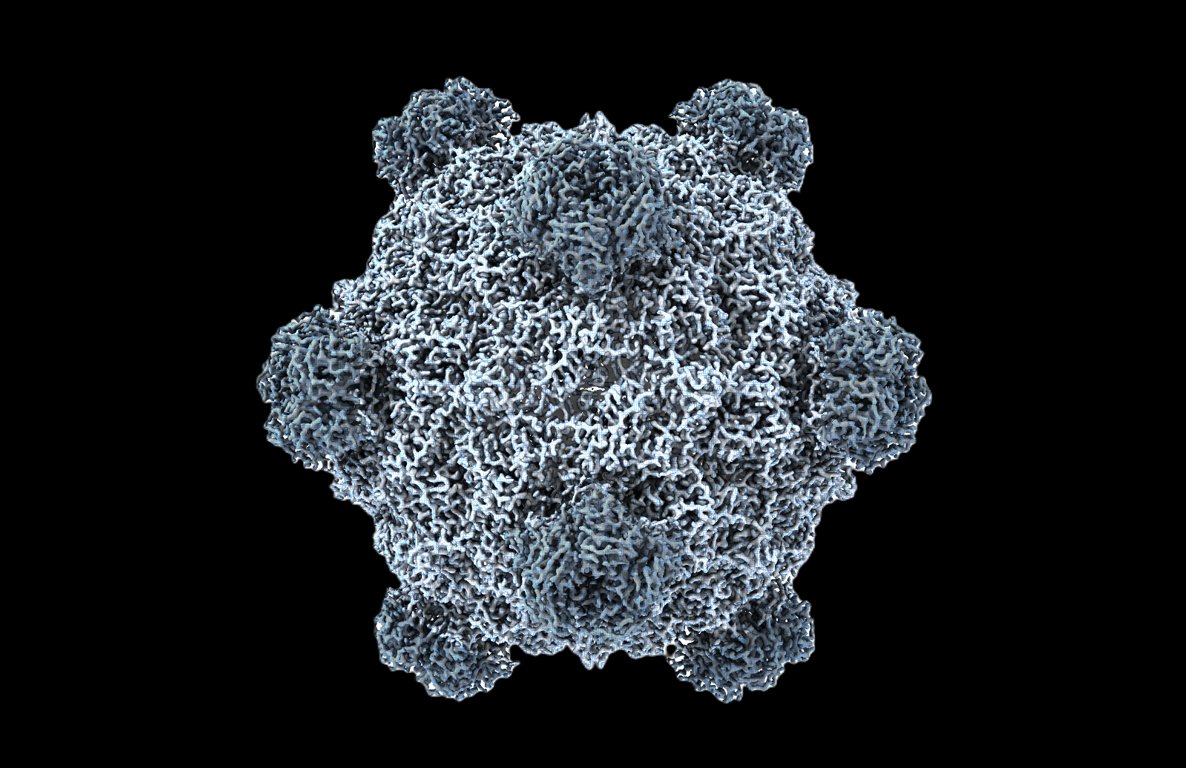
Welcome to the age of generative genome design! In 1977, Sanger et al. sequenced the first genome—of phage ΦX174. Today, led by Samuel King, we report the first AI-generated genomes. Using ΦX174 as a template, we made novel, high-fitness phages with genome language models. 🧵




🧬 Building the Virtual Cell starts with data. Today, we’re making the X-Atlas/Orion Perturb-seq dataset even more accessible — now live on Hugging Face Hugging Face 🤗 📊 One of the largest & highest-quality perturbation datasets ever released, it provides the foundation for