Naveen Jain
@uffdaallberries
Future physician-scientist in internal medicine and incoming intern @UCSF | @Penn ‘25 | @WashU ‘16 | functional genomics, lineage tracing, rare cell biology
ID: 1173970888007438342
17-09-2019 14:48:43
596 Tweet
300 Takipçi
499 Takip Edilen

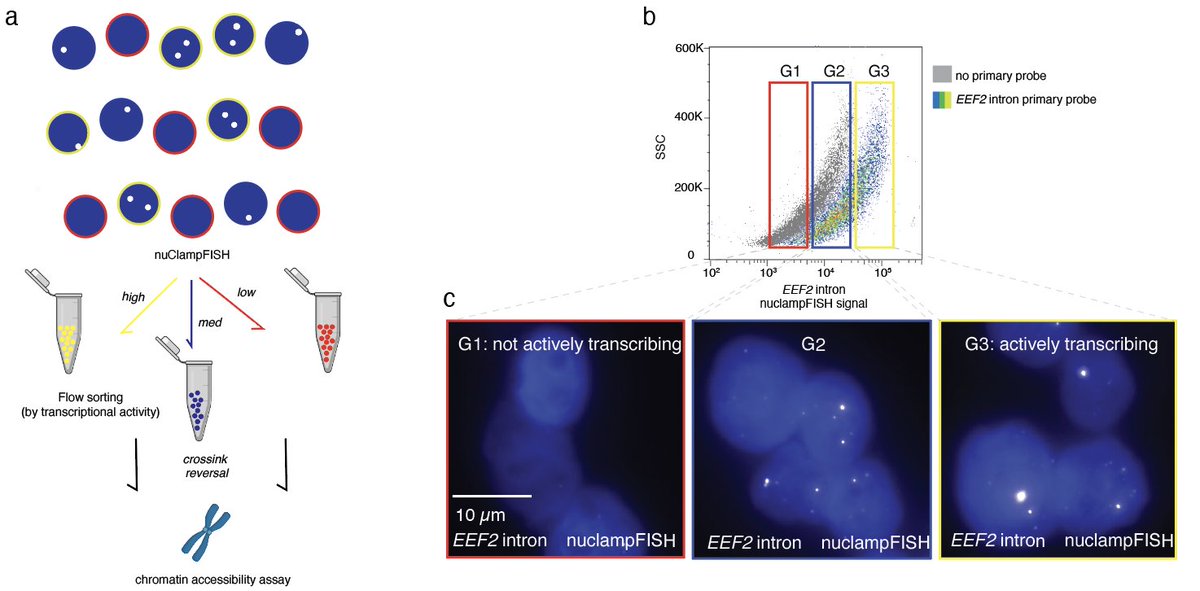
1/14 On behalf of the amazing team in Jonathan Weissman's Lab, we’re excited to share PEtracer (biorxiv.org/content/10.110…) a prime editing-based evolving lineage recorder compatible with both scRNA-seq and high-resolution imaging readouts in intact tissue. By applying PEtracer in a syngeneic mouse




New research out in Science Magazine provides a lineage-resolved analysis of embryonic gene expression evolution in C. elegans & C. briggsae ft. Christopher Large, Rupa Khanal, John Murray (Penn Genetics) & Junhyong Kim (Penn Biology) science.org/doi/10.1126/sc…



Very excited to share our new work on gastruloids by the incredible Cat Triandafillou! We mapped gene expression across 26 individual gastruloids at single-cell resolution and discovered some pretty amazing patterns about how these "mini-embryos" organize themselves.




I am a strong believer that depth of data can be just as powerful as scale of data. Deep analysis of small groups of humans, and even individuals, can lead to novel biological insights and principles. Today I am the guinea pig. In my own blood, Caleb Lareau finds strong evidence







Now online in Cancer Discovery: Systematic Evaluation of GAPs and GEFs Identifies a Targetable Dependency for Hematopoietic Malignancies - by Pu Zhang, Zhendong (Tom) Cao, Anthony Daniyan, Abdel-Wahab Lab, Junwei Shi, and colleagues doi.org/10.1158/2159-8… Penn Medicine - Abramson Cancer Center Memorial Sloan Kettering Cancer Center


Excited to share our CITE-seq atlas of the aging human immune system across tissues. Great collaboration with Donna Farber, Yosef Lab, Teichlab, Jo Jones Cambridge University and partnership with CZI Science! nature.com/articles/s4159…