Don’t miss Oxford Nanopore exhibiting at The Festival of Genomics & Biodata in Boston in June! Grab your ticket here: hubs.la/Q02ty0Hl0 #FOGBoston


📷📷📷📷
The deadline for workshops' pre-registration is Sunday, April 28. Registration to the meeting is FREE for attendees of the workshops! Visit hennigmeetingmexico.com/workshops.php
#Nanopore #NGS #SpeciesDiscovery #TNT #GeometricMorphometrics #STEM #Cladistics #Workshops #Science




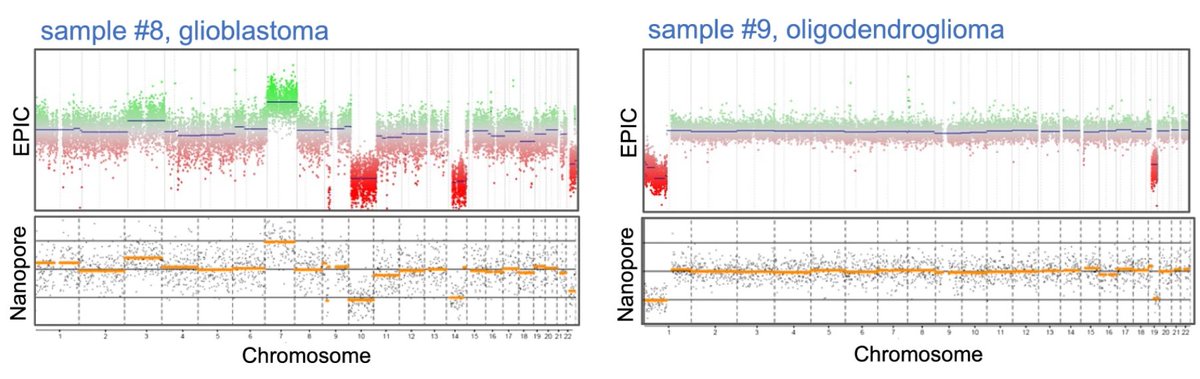
Always thought #Nanopore sequencing from #FFPE is impossible? Explore our paper: tinyurl.com/ffpe-nanopore. Methylation & CNV profiling of CNS tumors. Great collaboration with Ulrich Schüller and Leo. Oxford Nanopore

Delighted to launch this programme which enables Oxford Nanopore to continue improving genomics sequencing workflows for our end users via close collaboration with our partners. #NanoporeCommunity 📣

Ready for >Q30 ONT reads? Herro error correction now supports both R9.4.1 and R10.4.1. simplex reads w/ Dominik Stanojevic Dehui Lin (CHM13 -R9.4.1) #nanopore #longreads


“Characterising the synovial microbiome: a role for Nanopore sequencing”
Poster:P3545
Session 9b 30th April 12pm
Please come & say hello to our poster!
A collaborative effort with plastics, microbiology & the Hardman lab Hull York Medical School Hull University Teaching Hospitals NHS Trust SHYPS #eccmid2024

Happening now: Join the Chromatin Club Bay Area hosted by Active Motif to watch Ph.D. Candidate Colette Felton from the Brooks' Lab present her novel computational tool which uses nanopore data to identify gene fusions. Register here: activemotif.com/chromatin-club…


SimReadUntil for Benchmarking Selective Sequencing Algorithms on ONT Devices. #DeviceSimulator Oxford Nanopore #Bioinformatics
academic.oup.com/bioinformatics…

Have a read on my new piece on #biomarker discovery in #oncology for #pharma and #biotech in
rdworldonline.com/for-drug-disco…
Note the value of Oxford Nanopore flexible platform: short and long reads as well epigenetic information included.
What you are missing matters!



Drastic improvement for Oxford Nanopore DRS using kits RNA004 vs RNA002, confirming a similar observation previously by other labs Alexander Wittenberg Miten Jain. Average base error rate reduced from 10.2% (RNA002) to 4.2% (RNA004).



HiFi long reads outshine nanopore reads, delivering more insights with less sequencing.
Explore more about HiFi myths in cancer genomics research: bit.ly/3UkAJD8
#HiFiFacts


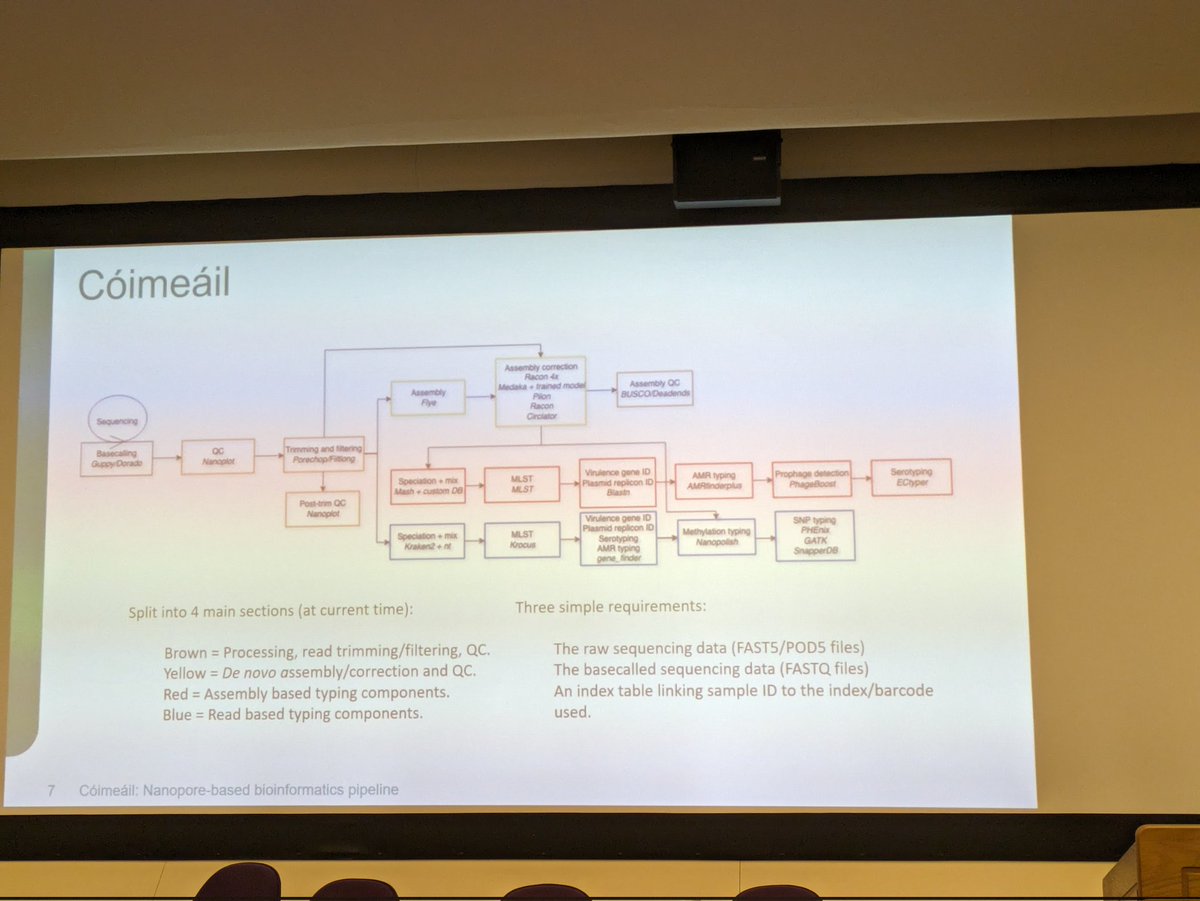
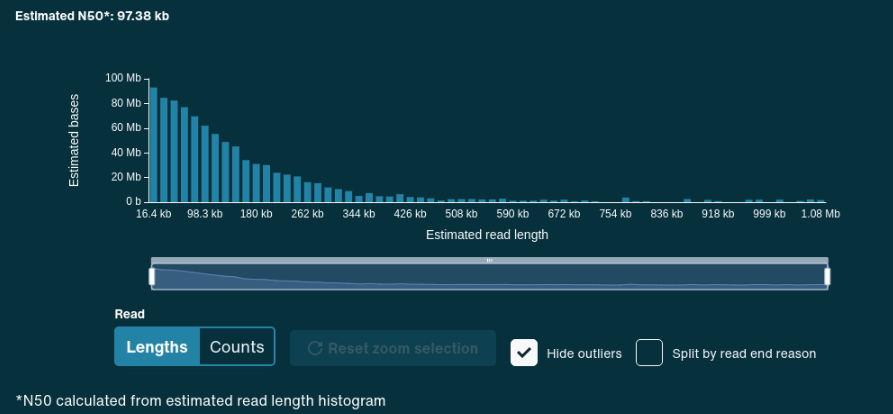
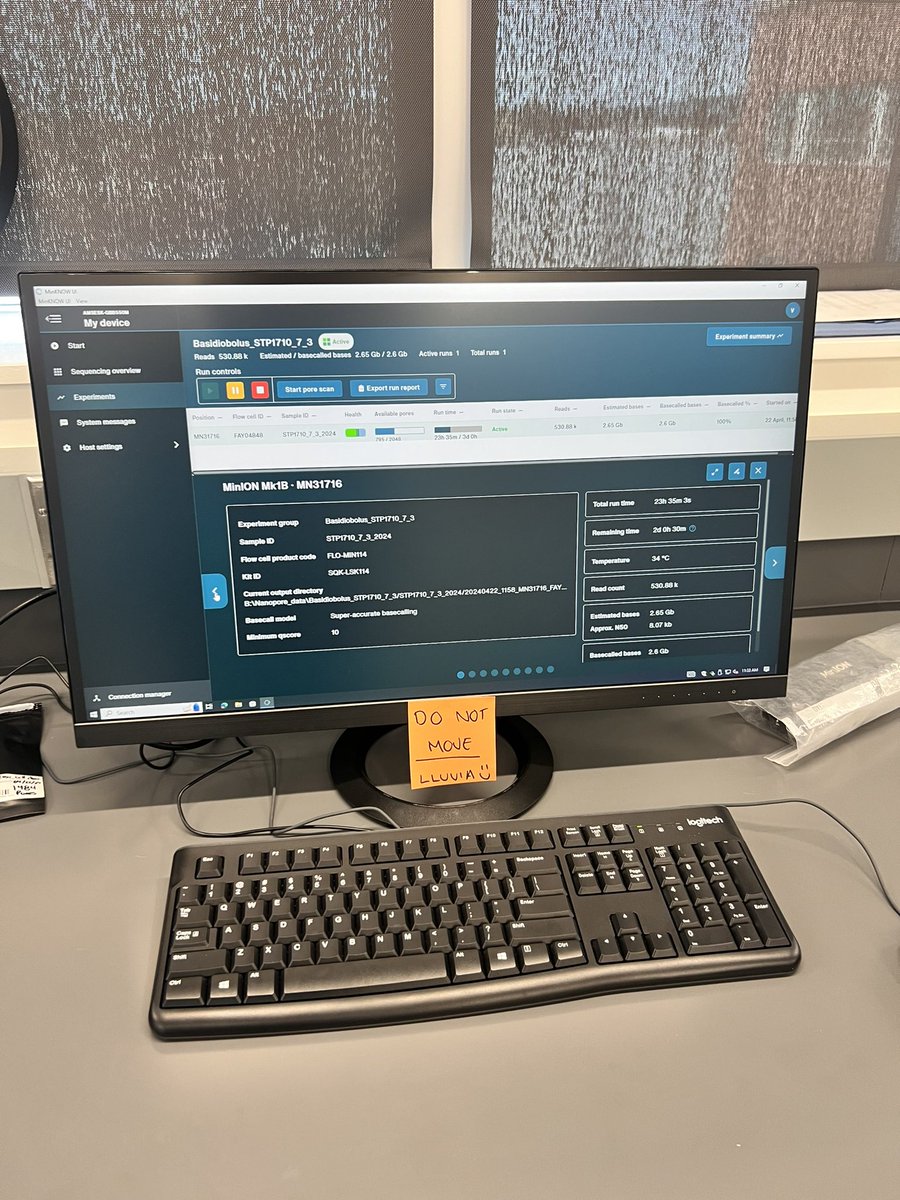
Here we go for the presentation of Cóimeáil the Nanopore based pipeline developed by David Greig from UK Health Security Agency
Extremely proud of the amount of work that went into it!
#hpruged HPRUged HPRU GI