Day 11 of #ADSB2024 , the Tiny Earth inspired short course on Antibiotic discovery Bioscience and Bioengineering IIT Jodhpur with Prof. Animalcules
We got about 200 Mb of data per isolate from our Oxford Nanopore MinION run after 20 hours. We assembled in Indian Institute of Technology Jodhpur computer center for data analysis.


Happy to present banana 🍌 pangenome using Oxford Nanopore duplex data at #nanoporeconf The insights obtained help in breeding new varieties that are resistant to Fusarium Tropical Race 4 (TR4) and Black Sigatoka. Thanks Willem van Rengs for the assemblies and comparisons.

-
a VERY RARE Capture of
The TheLongReadMaster
…. probably riding his horse 🐴
somewhere in West-Brabant …
Thank you Oxford Nanopore !


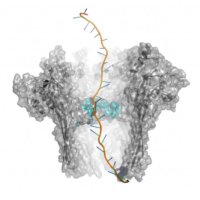
#SyracuseUniversity patent app, #US20240125728A1 for nanopore sensor with unprecedented specificity and single-molecule precision. This innovative design uses a protein pore and antibody-mimetic binder, capable of detecting diverse proteins by their unique electrical signatures
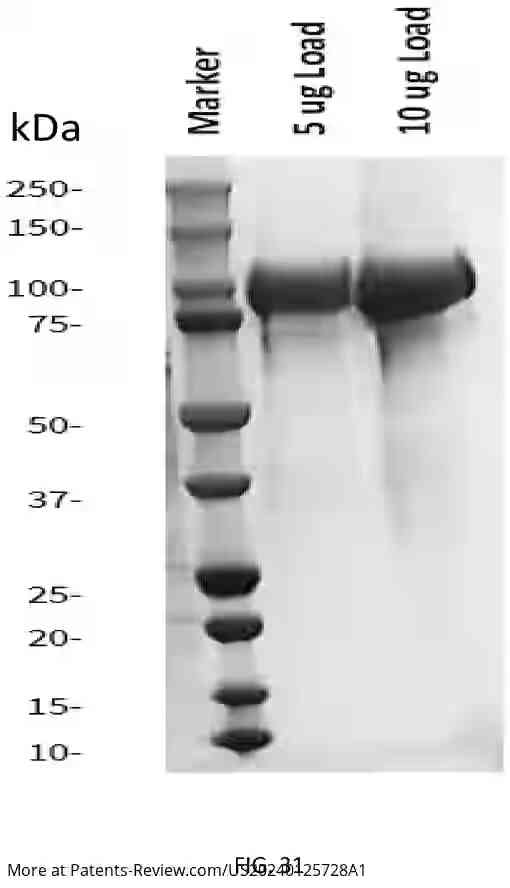

#LatePost
Day 10 of #ADSB2024 , the Tiny Earth inspired short course on Antibiotic discovery from soil bacteria Bioscience and Bioengineering IIT Jodhpur Indian Institute of Technology Jodhpur with Animalcules
As our Oxford Nanopore MinION was spewing out reads, we prepared for genome analysis with a lecture on microbial genomics.

It begins #ESCMID2024 ! Swing by booth C3 to meet the Oxford Nanopore team & discuss infectious disease applications including:
🧫 Microbial isolate whole genome sequencing
🦠 Full-length 16S microbial identification
🤧 Influenza WGS
🧬 Pathogen-agnostic metagenomics
#ESCMIDGlobal


🍿 Pop corn ✅
🪩 Party ✅ thank you Oxford Nanopore
🥰 Perfect company ✅
🦠 Part 3 of #ESCMIDGlobal2024 here we go!


Just got back after a fantastic 2 days at the #UKDNAWG24 conference where I presented my PhD research - using Oxford Nanopore Flongles for #DNA barcoding and #eDNA #metabarcoding . What a wonderful community, excited for next year already!





Discover how multiomic nanopore sequencing on a single platform can be used to maximize the data generated from your valuable research samples in Oxford Nanopore’s latest Brochure.
Download now to learn more >>> hubs.ly/Q02tT-pt0


Register here - lnkd.in/daXyJRvcLive reviews - lnkd.in/d6a3_Z4H #BasicsOfNGS #Illumina #IonTorrent #Nanopore #DNASeq #PacBio #NGS_data_analysis #Anaconda #Conda #Conda #SamTools #bcftools #fastqc #multiqc #BurrowsWheelerAlignmentAlgorithm #Bowtie #DrVipinsWorkshop