Nancy Yu
@ynancy
Genomics, bioinformatics and health/medicine - related (re)tweets
ID: 18228351
18-12-2008 23:12:42
4,4K Tweet
416 Followers
1,1K Following

🚨preprint alert🚨 We report a #GWAS with ratios between protein levels from the Olink Proteomics Explore 1536 #proteomics data from ~54,000 participants of the UK Biobank using the DNAnexus, Inc. platform. biorxiv.org/content/10.110… 1/13

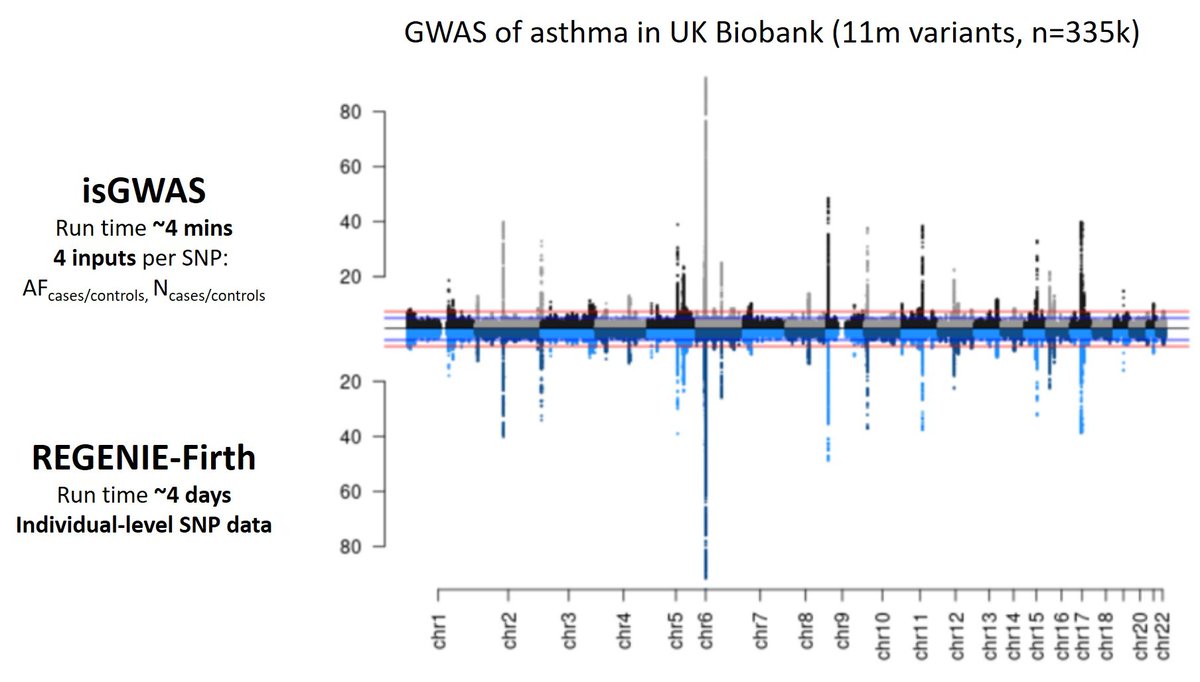
Case-control GWAS on 11m variants in 335k people in <4 mins using ONLY summary-level data! Check out our isGWAS bioRxiv at biorxiv.org/content/10.110… - large multi GWAS now accessible to all! Made possible with MarioniGroup Heiko Runz Zhana & Chris Foley!

Proteomics outshines polygenic risk scores in predicting 218 common and rare diseases. Impressive C-statistics for some diseases for which we have few predictive tools in the clinic. omicscience on a roll with Robert Scott @pietznerm and others medrxiv.org/content/10.110…

Anyone who wants to study #HLA but feels hesitant as HLA is too complicated☹️? Happy to share our paper Nature Protocols describing the up-to-date knowledge on HLA #genetics and step-by-step tutorial for HLA imputation and association with Soumya Raychaudhuri সৌম্য রায়চৌধুরী🥳 nature.com/articles/s4159…

A genome-wide meta-analysis of data from 6 US and European cohorts involving 1.3 million individuals identifies 243 genetic variants associated with risk and pathophysiology of depression Anders Børglum nature.com/articles/s4159…


New research from Sergio Villicaña Jordana Bell & co. King's College London, with findings that improve the characterisation of the mechanisms underlying DNA methylation variability & are informative for prioritisation of GWAS variants for functional follow-ups bit.ly/3qh5pJi
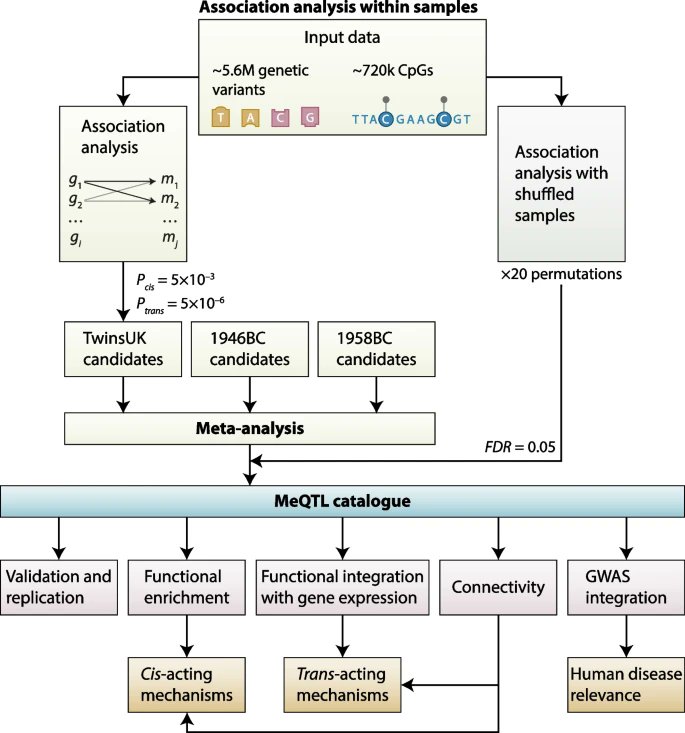

This is as close as our group could get to a how-to guide for studies of the environment and #epigenetics Don't forget DNA sequence variation. Look for transcription factor and cell signalling effects. @_jjdubs David A Knowles (@davidaknowles.bsky.social) Sri Raj journals.plos.org/plosgenetics/a…




Excited to share our work introducing a remarkably efficient approach to model linkage disequilibrium, now published in Nature Genetics nature.com/articles/s4158… This work was led by Pouria Salehi Nowbandegani and @WilderWohns

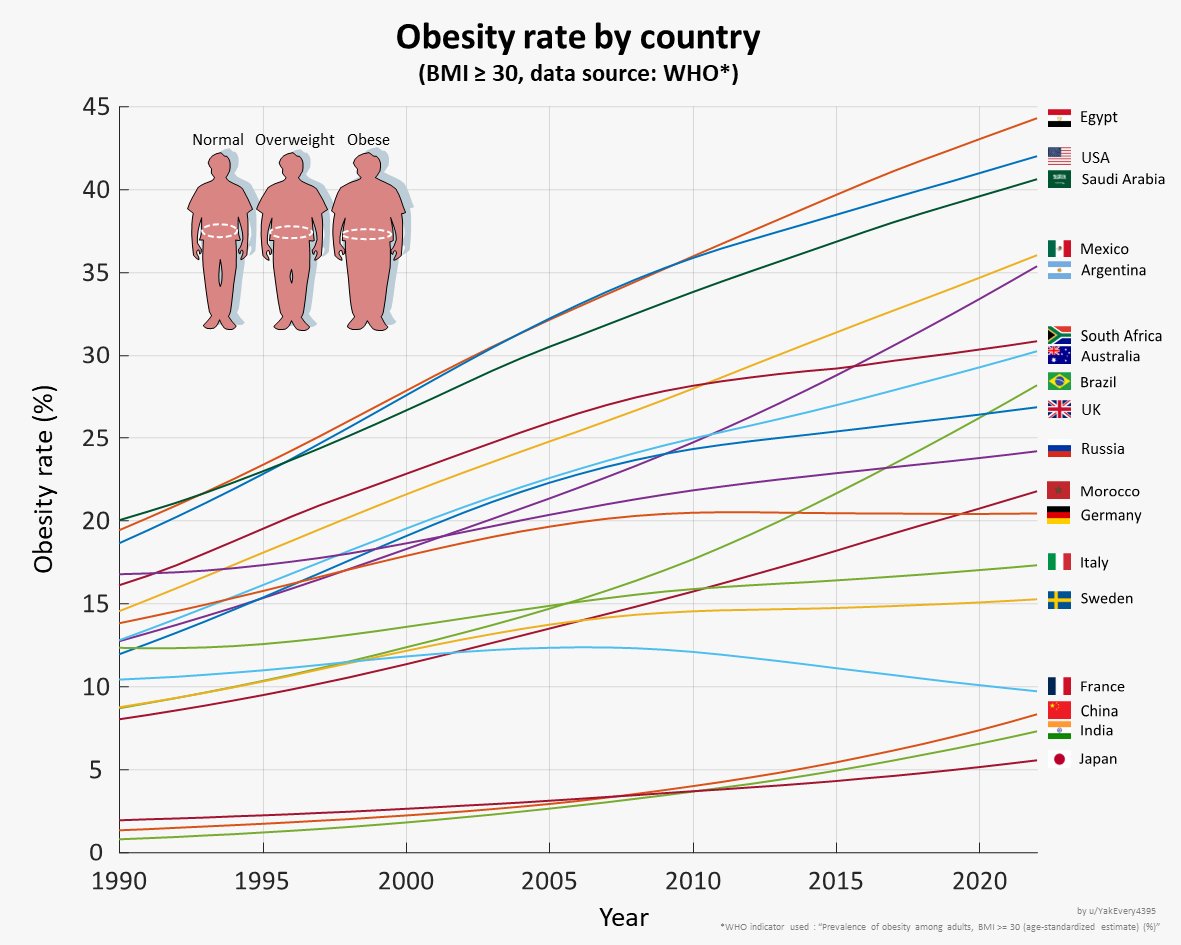
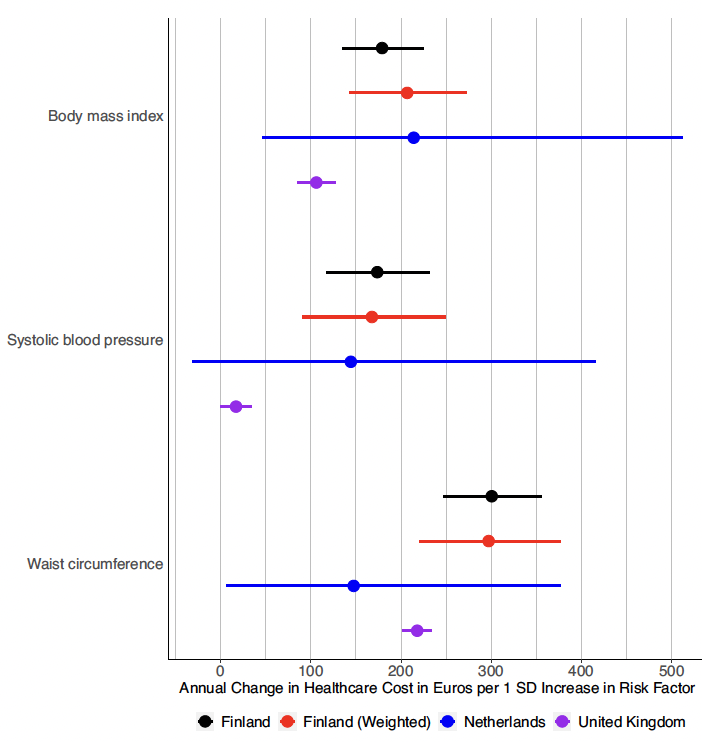
Huge congrats to our HST Harvard Medical School student Jiwoo Lee using Mendelian randomization to estimate healthcare costs attributed to various clinical risk factors. Highlights substantial estimated cost savings from treating obesity nature.com/articles/s4146… Nature Communications Andrea ganna




Very happy to present our new paper led by an excellent team of graduate students Michael Slobodyanyuk Alec Bahcheli at Ontario Institute for Cancer Research (OICR) Department of Molecular Genetics, U of T Medical Biophysics: nature.com/articles/s4146… 1/4