
Ruzhang Zhao
@ruzhangzhao
PhD student in Biostatistics at Johns Hopkins Univ.
BS in Mathematics from Tsinghua Univ.
ID: 1112690153170960384
http://ruzhangzhao.com 01-04-2019 12:16:20
51 Tweet
162 Followers
301 Following

A year ago in Nature Biotechnology, Becht et al. argued that UMAP preserved global structure better than t-SNE. Now George C. Linderman and me wrote a comment saying that their results were entirely due to the different initialization choices: biorxiv.org/content/10.110…. Thread. (1/n)




Fun fact: There are about 2,700 trees on the Johns Hopkins University campus, not including forest areas. (📷 Will Kirk / Johns Hopkins University)


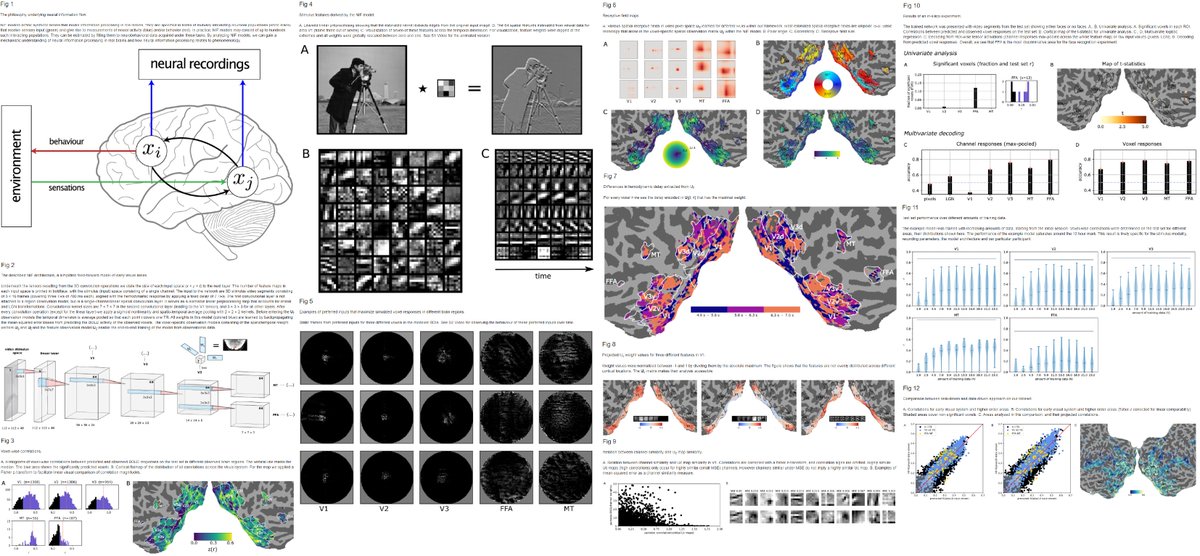
"End-to-end neural system identification with neural information flow". Our new PLOS Comp Biol methods paper has been published. doi.org/10.1371/journa… w/ @kateiyas, Luca Ambrogioni, Dr. Yağmur Güçlütürk & Marcel van Gerven. #fmri #systemidentification #visualsystem

Excited to share our latest manuscript on complex trait fine-mapping across diverse populations! This is a collaborative effort across BioBank Japan FinnGen and UK Biobank with mentorship from Hilary Finucane Mark Daly YukinoriOkada. Here is 🧵 1/n medrxiv.org/content/10.110…

Happy to share our new preprint on meta-analysis fine-mapping in the Global Biobank Meta-analysis Initiative Global Biobank Meta-analysis Initiative. Joint work w/ Roy Elzur Wei Zhou under the mentorship of Mark Daly Hilary Finucane. Tl;dr meta-analysis fine-mapping is hard! 1/n medrxiv.org/content/10.110…



New GWAS of T2D (180K cases, 1.2M controls; ~50% non-Eur). Fine-mapping substantially improved with non-Eur samples prioritizing single causal variants at 54% of the 237 loci. Nature Genetics




A very nice collaboration on multi-ancestry PRS with a combination of penalization and ensemble learning methods. Great collaborations with Jingning Zhang Nilanjan Chatterjee Jin Jin Jianan Zhan Ruzhang Zhao 23andMe @NCIEpiTraining and all others colleagues!

New paper from our lab on methods for bulding high-dimensional predictive models with disparate datasets. Amazing team work by Ruzhang Zhao Prosenjit Kundu and Arkajyoti Saha. arxiv.org/abs/2312.12786 1/5


Thanks for the amazing mentorship of Nilanjan Chatterjee and collaboration with Prosenjit Kundu Arkajyoti Saha !

Our paper on MUSSEL, a method for developing ancestry-specific PRS, is finally out in Cell Genomics : cell.com/cell-genomics/…! Thank you for the > 3-year effort Nilanjan Chatterjee Jianan Zhan Haoyu Zhang Ruzhang Zhao Updated software: github.com/Jin93/MUSSEL Feel free to try it out!

Our paper on PROSPER, a new multi-ancestry PRS method with penalized regression is out on Nature Communications ! Check here nature.com/articles/s4146…. Thanks to everyone offering help along the way!! Nilanjan Chatterjee Haoyu Zhang Jin Jin Jianan Zhan Ruzhang Zhao Cheng Ma and PRIMED Consortium







