YukinoriOkada
@okada_yukinori
Professor, Dep. Genome Informatics, The University of Tokyo
Professor, Dep. Statistical Genetics, Osaka University
Team Leader, Lab. Systems Genetics, RIKEN IMS
ID:1175322065815031809
http://www.sg.med.osaka-u.ac.jp/index.html 21-09-2019 08:13:22
220 Tweets
2,0K Followers
812 Following


Today, we discussed about a paper by YoshihikoTomofuji and YukinoriOkada on multi-omics analysis of host genotypes, gut microbiome and plasma metabolome. The main idea is to detect gut microbiome-host interaction through metabolites activities. 1/2 doi.org/10.1016/j.celr…



Multi-omics network of Japanese (gut microbiome🦠 plasma metabolome🧪 host genome🧬) by YoshihikoTomofuji is now Cell Reports㊗️🎉.
ABO blood type🆎 associate with gut microbial genes related to N-galactosamine metabolism.
cell.com/cell-reports/f…
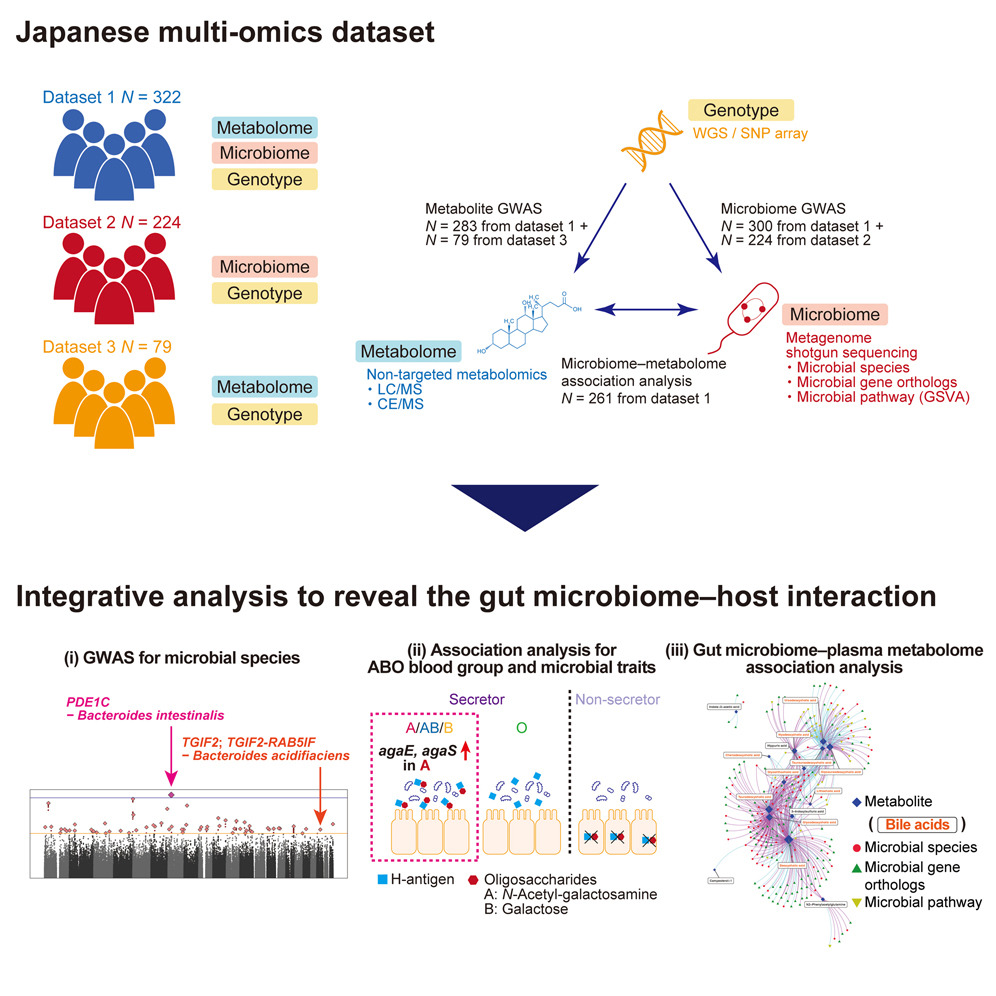

We are happy to announce that our multi-omics analysis of the gut microbiome (gut microbiome + human genotype + plasma metabolites) is finally published from Cell Reports (cell.com/cell-reports/f…)🎉🎉🎉! GWAS sumstats are available from NBDC (humandbs.biosciencedbc.jp/en/hum0197-v18…).

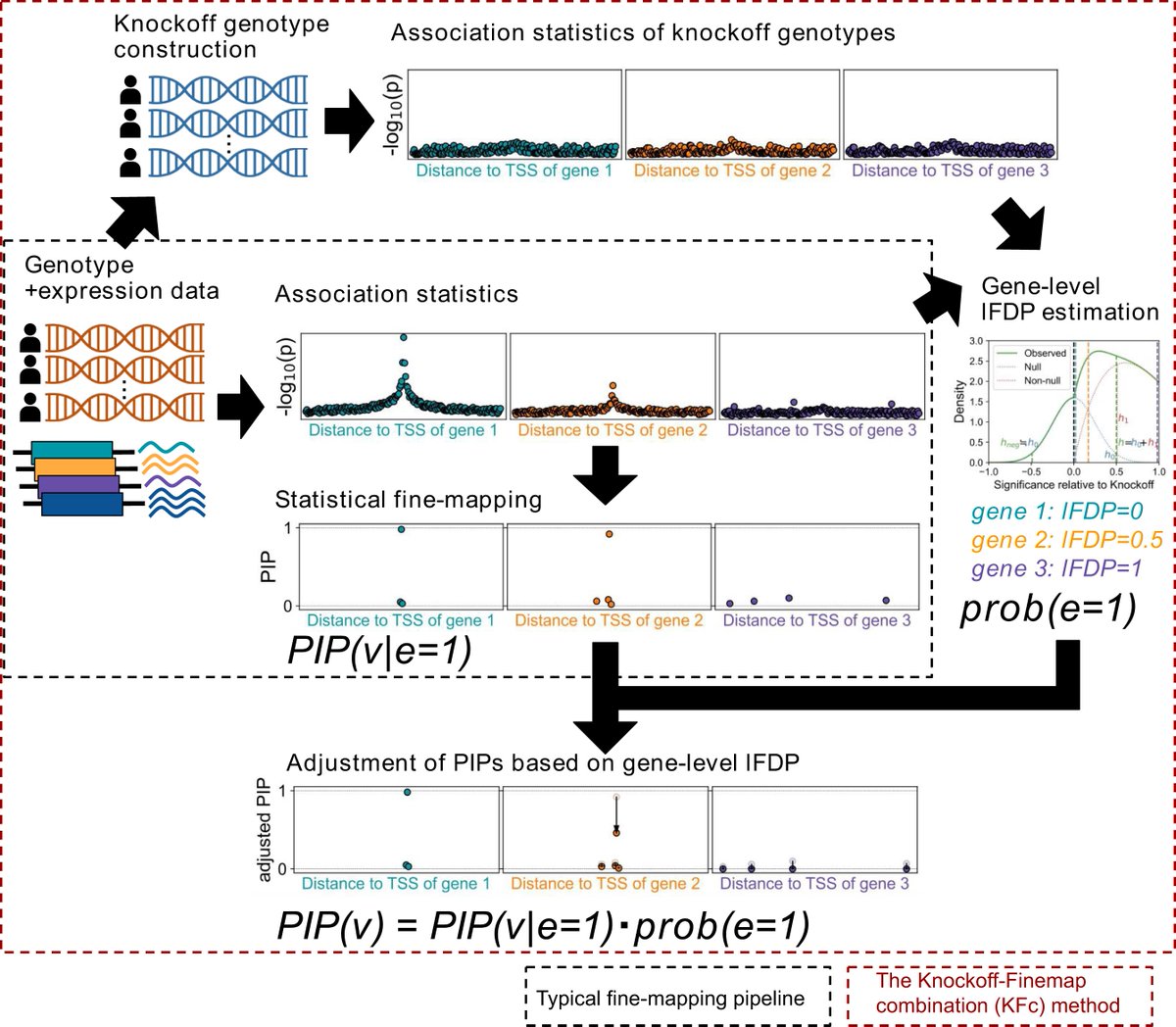
New application of knockoff genotypes (synthetic genotypes resembling original ones) for omics resources by Qingbo Seiha Wang. Knockoff–Finemap combination (KFc), a refined fine-mapping algorithm of eQTL to adjust PIPs. Now at NAR Genom Bioinform🎉㊗️
academic.oup.com/nargab/article…




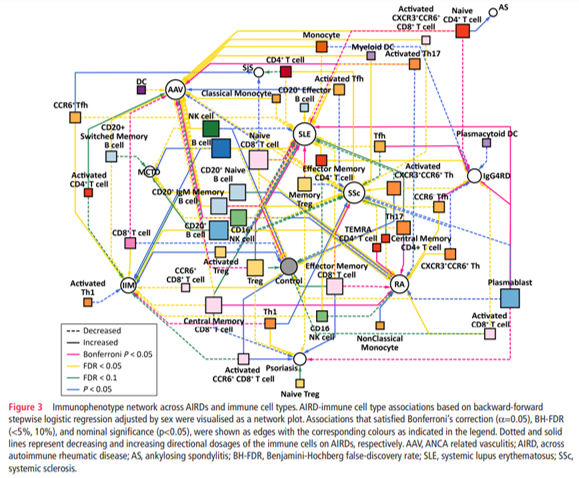
🆕 #immunophenotyping of 46 peripheral immune cells in >1000 Japanese patients of 11 AIRDs 🗾
🎭 RA-like or SLE-like clusters were exclusively dominant
👉 clinically heterogeneous patient subtypes exist within single disease
🔗 bit.ly/49iHgnp
YukinoriOkada


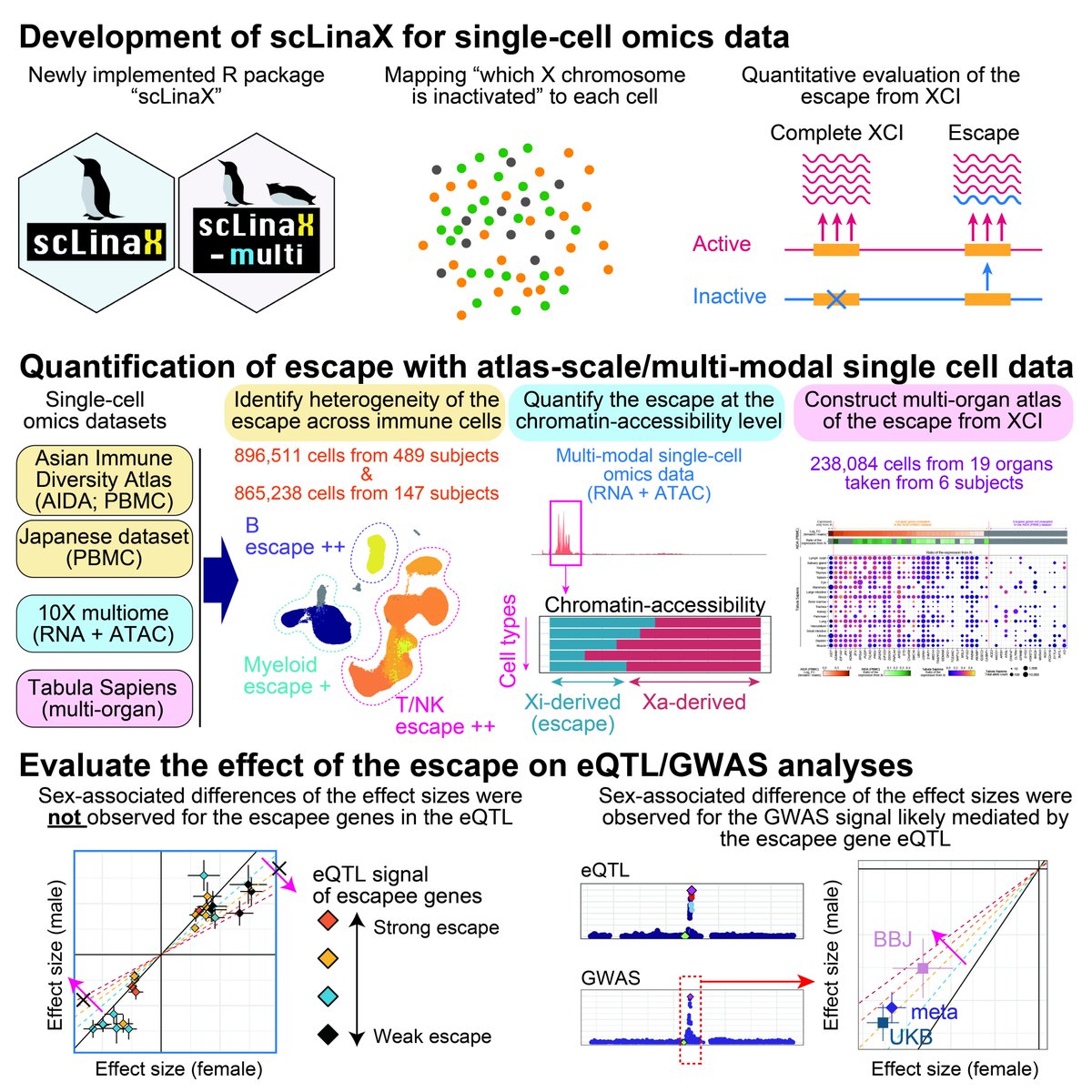
Half or X chromosomes in females are inactivated (XCI), but some escape. scLinax: tool to quantify XCI escape using millions of single cells by YoshihikoTomofuji as Asian Immune Diverstiy (AIDA) Nework projects of Human Cell Atlas with Shyam Prabhakar Lab & colleagues. biorxiv.org/content/10.110…