
Anurag Vaidya
@anurag_vaidya7
Machine learning and healthcare enthusiast
ID: 1515345326823063556
16-04-2022 15:04:33
36 Tweet
68 Followers
150 Following

1/4 Interested in multimodal fusion in Computational Pathology? Checkout our preprint combining transcriptomics and histology for survival prediction! Paper: arxiv.org/abs/2304.06819 Code: github.com/ajv012/SurvPath Anurag Vaidya Richard J. Chen @DFKW_MD Paul Liang Faisal Mahmood


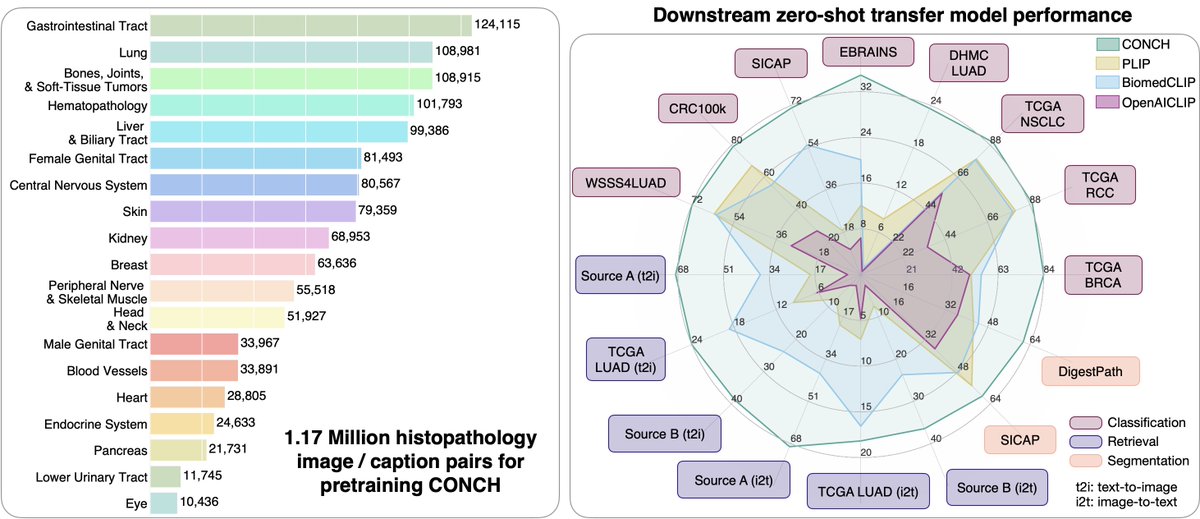
⚡️🔬📣Excited to share our two new Nature Medicine articles, we develop computational pathology foundation models, 1. UNI, a self-supervised computational pathology model trained on 100 million pathology images from 100k+ slides. 2. CONCH, a vision-language model for

.Raj Movva, Pang Wei Koh, and I write for Nature Medicine on using unlabeled data to improve generalization + fairness of medical AI models: nature.com/articles/s4159… We highlight two nice recent papers illustrating this - nature.com/articles/s4159…, nature.com/articles/s4159….

⚡️🔬📣Excited to share our new Nature Medicine article, examining disparities in pathology AI models, assessing how modeling choices impact disparities, and evaluating the potential of self-supervised foundation models in mitigating these disparities. nature.com/articles/s4159… See



Excited to share our latest publication in Nature Medicine! 🎉 Proud to have been part of this incredible team effort. We study the disparity and fairness in AI models for computational pathology, exploring a variety of modeling strategies. Check it out below! 👇

What an incredible year for Faisal Mahmood lab! Leading the field in computational pathology and now doing excellent work on disparities.

New studies suggest that using unlabelled data in medical #AI can improve accuracy and generalization to new settings and minority patient groups, thereby increasing fairness. News and Views from Emma Pierson & colleagues Emma Pierson Cornell Tech nature.com/articles/s4159…



Based on numerous requests we are providing the open access ShareIT link for our Nature Medicine article on identifying and mitigating disparities in pathology AI models. Open read link: rdcu.be/dFdMS Journal link: nature.com/articles/s4159…

Excited to share our review on hemostatic agents, where we summarize trends in 54 approved hemostats and 75 active clinical trials. Check out this timely article - with Pfizer’s latest gene therapy for Hemophilia B approved just last week! Samir Mitragotri Zongmin Zhao @BioTM_Buzz



Using artificial intelligence, researchers from Mass General Brigham have created and trained a new model using 3D datasets to predict the recurrence of prostate cancer. The model outperformed other models that rely on 2D datasets. buff.ly/3VnW7YH Faisal Mahmood Andrew H. Song


🚨⚡️Super excited to be part of HEST-1K, bringing you over 1.5M histology-omics pairs. Have fun with this public dataset for representation learning, biomarker discovery, and benchmarking models! Massive efforts by Guillaume Jaume Paul Doucet Faisal Mahmood

🚨⚡️Excited to bring you MMP - a highly interpretable and computationally light method to do multimodal prognostication in computational pathology. Amazing effort by Andrew H. Song Richard J. Chen Guillaume Jaume and Faisal Mahmood

🚨🔬 Super thrilled to announce a fun project with Guillaume Jaume! Using 7,000+ slides and 20+ tasks, we show that aligning H&E and immunohistochemistry slides results in robust gigapixel slide encoders.





