Antonio Rios
@antonio_science
Aviv Regev’s Lab @Genentech | previously Engreitz Lab @Stanford | @GatesFoundation Scholar | applying to bioe/genetics PhD programs for Fall 2025
ID: 1269821233702424576
08-06-2020 02:39:18
51 Tweet
156 Followers
497 Following


Our preprint on the development of PerturbView, an Optical Pooled Screening (OPS) technology, is out! Massive thanks to Eric Lubeck for driving the project with me, along with our fantastic mentors, Orit Rozenblatt-Rosen, Levi A. Garraway and Aviv Regev biorxiv.org/content/10.110… 1/


Spatial analysis is converging with pooled CRISPR screens in a recent series of preprints w/ different guide readouts: 1. RCA biorxiv.org/content/10.110… 2. in situ txn + FISH biorxiv.org/content/10.110… 3. in situ txn + in situ seq biorxiv.org/content/10.110… Exciting for tissue biology!






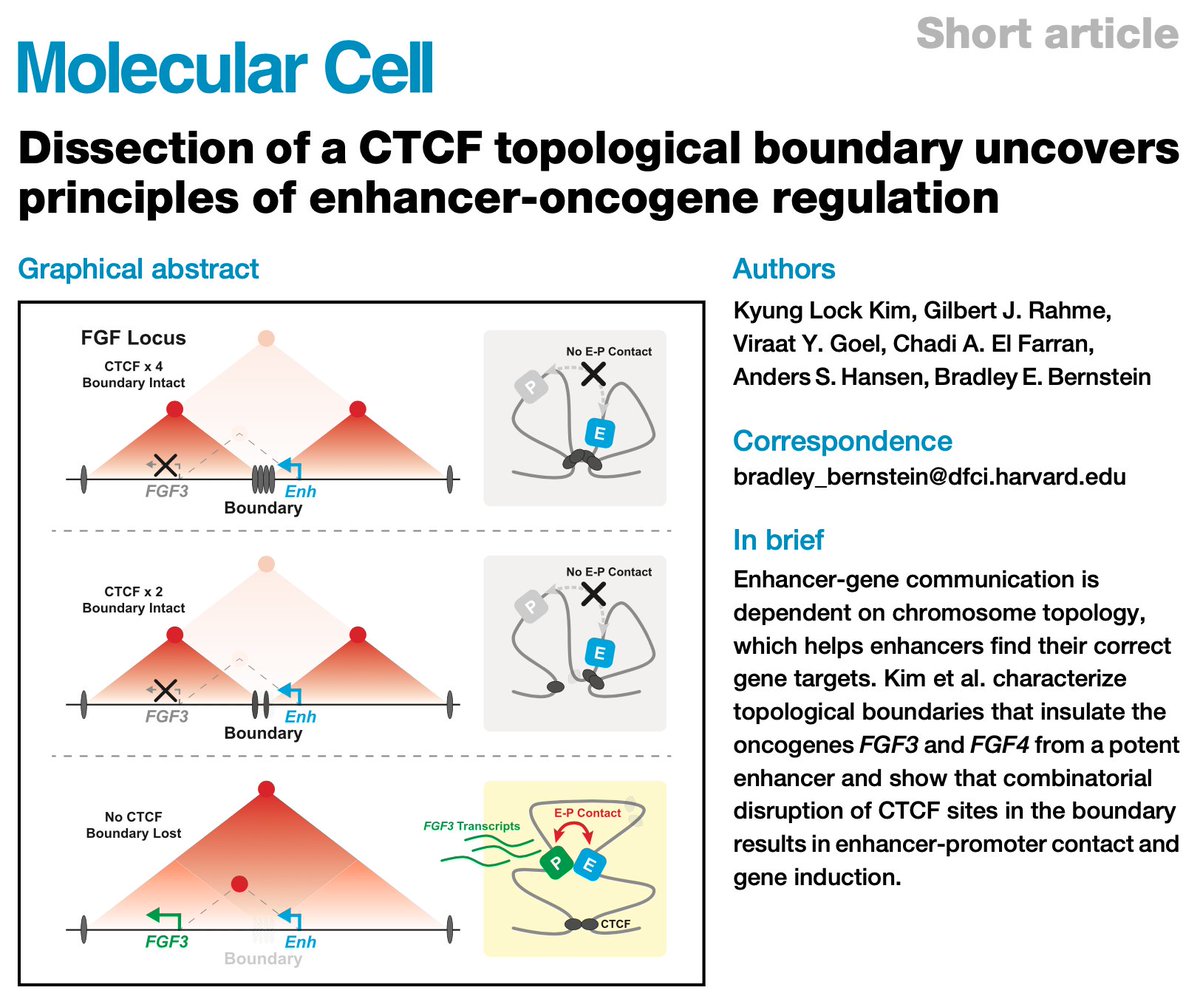
(1/2) New paper from KL Kim Gilbert J. Rahme (He/him/his) Brad Bernstein Gene Regulation Observatory with RCMC contribution from Viraat Goel ASHansen Lab sciencedirect.com/science/articl… Quantitative dissection of how loss of a CTCF insulator boundary in GIST causes aberrant enhancer-oncogene interactions



Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment. nature.com/articles/s4158…