Cuong To
@totucuong
#protein #ML #startup
ID: 42263193
https://totucuong.github.io/ 24-05-2009 19:11:01
25 Tweet
46 Takipçi
149 Takip Edilen

Did you miss the chance of joining the #GCSB2020 #SynBio #Conference? Here is a summary of the two days: bit.ly/30knoyq. Also, we will upload the videos of the talks soon to ga-sb.de. Stay tuned and see you next year at the #GCSB2021 Centre for Synthetic Biology @TUDarmstadt!


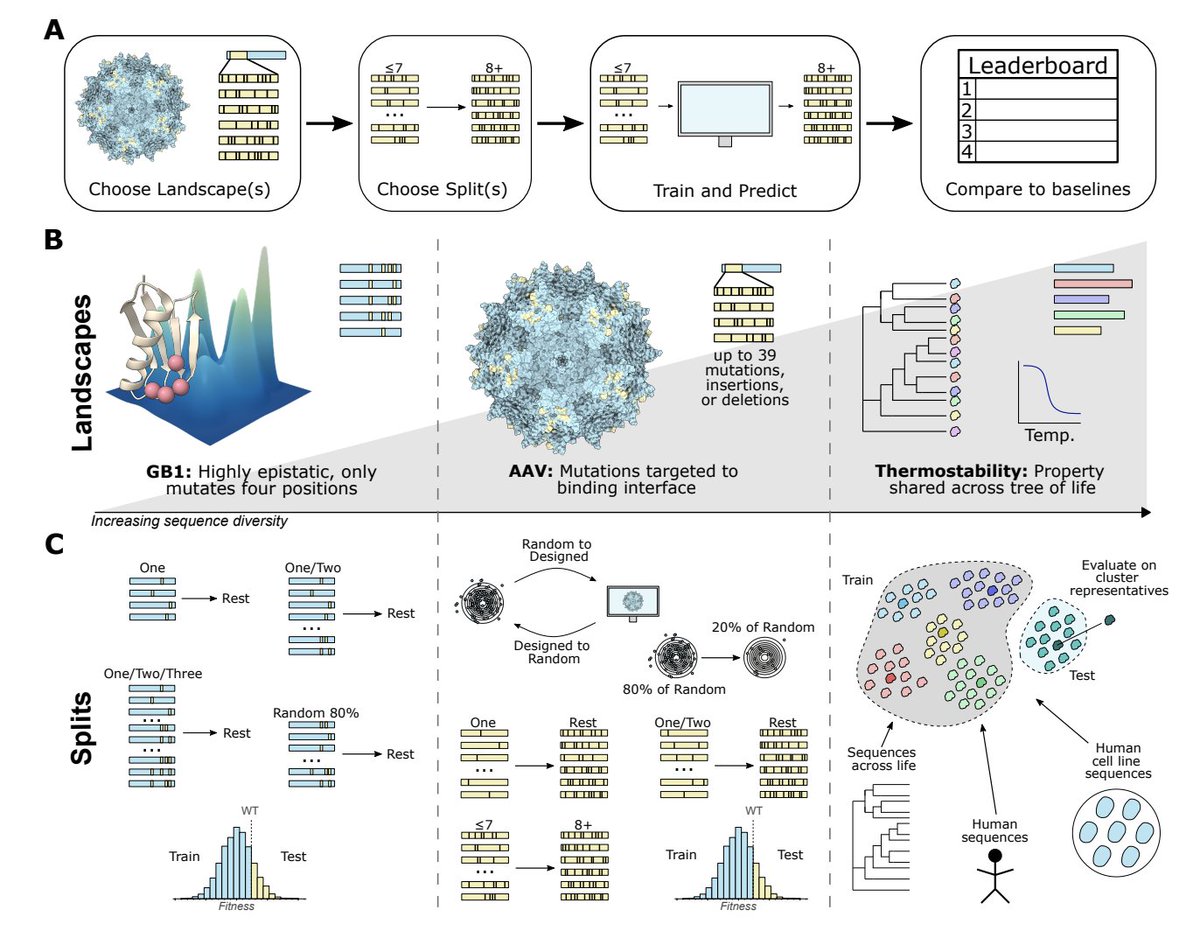
Testing the ability of different ML methods to generalize on synthetic and experimental protein landscapes. Adam C Mater, Mahakaran Sandhu, Colin Jackson biorxiv.org/content/10.110…



3/ Eric Kelsic (he/him) and I also gave a talk at the Broad on this subject which also covers how we approach AAV design Dyno Therapeutics, Inc. . The first half is a primer on ML-guided sequence design, and the second half covers our work on AAV design. youtu.be/QLURMsm72cE

#MaxSynBio2020 is taking place from Nov 30 to Dec 2. Hoping that a large and international bottom-up synthetic biology crowd can join us for this online meeting! maxsynbio.mpg.de/symposium-2020 MaxSynBio MaxPlanckBristol Synthetic Cell initiative Build-a-Cell GASB SynBio Registrations are open & free.


Can we predict protein structure directly from sequence w/o MSAs and w/o intermediate steps like distograms? In a new preprint we show that we can, led by Ratul Chowdhury Nazim Bouatta Surge Biswas along w/Charlotte Rochereau george church & Peter Sorger: biorxiv.org/content/10.110… (1/4)

Welcome if you're joining us for our Pacific Time session of Writing the Future of Drug Discovery and Development - we've got more great speakers coming up including Sai Reddy chris bahl Carterra Ginkgo Bioworks @barrettnuttall #writingthefuture2021 bit.ly/3AA8Fi1

A set of benchmark tasks for predicting protein fitness or function from sequence. Christian Dallago Jody Mou Kadina Johnston Bruce Wittmann, Nicholas Bhattacharya, Sam Goldman Ali Madani and me (Kevin K. Yang 楊凱筌) biorxiv.org/content/10.110…

An announcement I’ve been aching to make! After much sweat, we’ve built a trainable version of AlphaFold2, implemented in PyTorch, which we’re calling OpenFold. GitHub: github.com/aqlaboratory/o… Colab: colab.research.google.com/github/aqlabor… Why a trainable version of AlphaFold2 you ask? ⬇️


after a brief hiatus over the holidays, I'm excited to announce that we're back! We've got a great workshop by Sergey Ovchinnikov for you next Wednesday February 2nd at 7pm EST Sergey Ovchinnikov "Tutorial on using structure prediction methods for protein design" harvard.zoom.us/j/94214636314

Watching DE-STRESS (pragmaticproteindesign.bio.ed.ac.uk/de-stress/) presentation at #apfed22 . Weight&Bias for protein engineering 😍 Christopher W. Wood

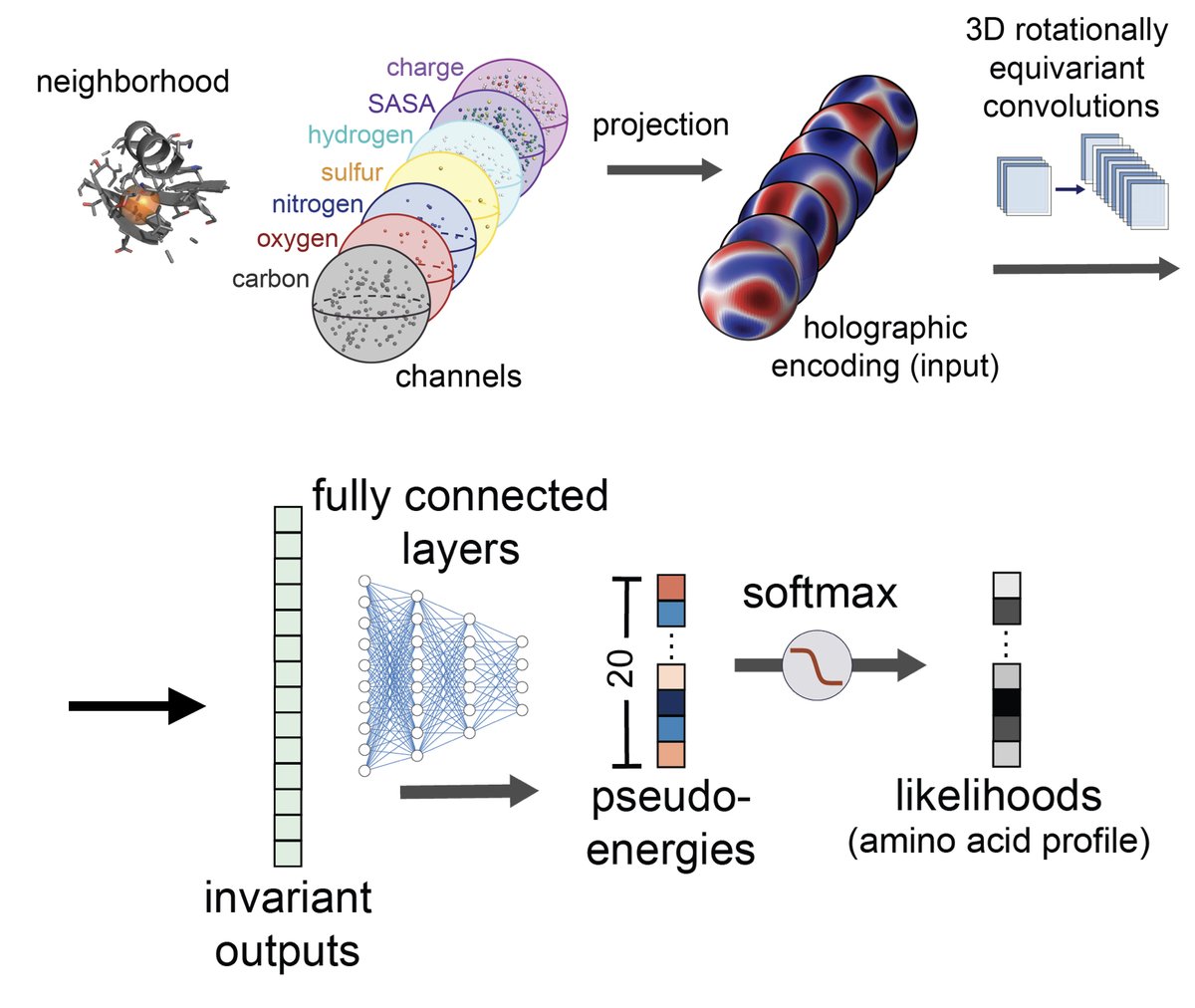
Check out our new preprint (led by Mike Pun University of Washington) on 3D rotationally equivariant model of protein structure micro-environments, with a holographic CNN (H-CNN): doi.org/10.1101/2022.1…