Tom Sercu
@tomsercu
Building @evoscaleai - Frontier AI for biology. Ex-Meta FAIR, Ex-IBM Research. Alum @NYU, @ugent.
ID: 506357248
http://tom.sercu.me 27-02-2012 20:28:30
357 Tweet
2,2K Followers
739 Following


We had an excellent demo and presentation from Roshan Rao of EvolutionaryScale at #LuxAISummit, which uses AI models (like ESM3) to assist scientists to understand, imagine, and create proteins.




Really exciting new series of highly efficient protein language models from EvolutionaryScale


Whoa! When a large language of life model generates a protein equivalent to ~500 million years of evolution. Science Magazine science.org/doi/10.1126/sc… Thomas Hayes Roshan Rao EvolutionaryScale Arc Institute UC Berkeley


Thrilled that the ESM3 paper is now published in Science Magazine! 🌐🧬 We are also announcing public beta availability of the models via the Forge API, including demos to generate proteins and predictions with ESM3 without need to write any code!

So excited about the progress by EvolutionaryScale with ESM3 now in Science Magazine, ESM Cambrian, and more to be announced. Thrilled to continue our collaboration!





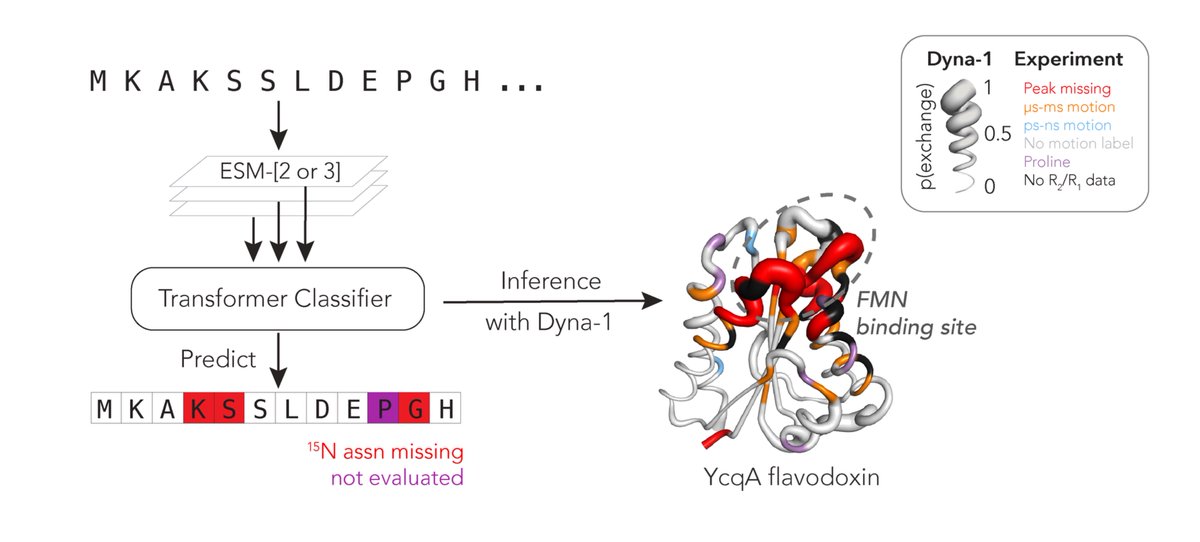
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵