
Grant
@synbiogrant
ID: 1238242001046421506
12-03-2020 23:14:45
412 Tweet
430 Followers
369 Following


Self-assembling nanoparticle technology created at King Lab is the basis for a #COVID19 vaccine approved in South Korea. Now, an #RSV vaccine with the same platform is in clinical trials through @icosavax. UW Medicine Institute for Protein Design UW Biochemistry bit.ly/3UMUfWQ

Are you ready to learn about the most exciting innovations in #synbio #SB? The 2023 SEED Conference was just announced from May 30-June 2 Mikhail Shapiro (same on bsky) 🇺🇦 Christopher Voigt Northwestern CSB + more synbioconference.org/2023

[PLEASE RT] With my awesome co-chairs, I'm excited to share the (nearly complete) program for one of my FAVORITE meetings: Synthetic Biology Gordon Research Conferences 2023. This meeting is special -- I highly encourage folks to register and attend! MicroDynamic 🌱 grc.org/synthetic-biol…





Interested in protein design, deep learning, computational biophysics? Or computational RNA folding, vaccines, enzymes, antibodies, biomaterials, machine learning? Become a Rosetta Commons undergraduate summer intern (U.S. National Science Foundation #REU) or a postbaccalaureate fellow (#RaMP)! [thread!]
![Jeffrey J. Gray (@jeffreyjgray) on Twitter photo Interested in protein design, deep learning, computational biophysics? Or computational RNA folding, vaccines, enzymes, antibodies, biomaterials, machine learning? Become a <a href="/RosettaCommons/">Rosetta Commons</a> undergraduate summer intern (<a href="/NSF/">U.S. National Science Foundation</a> #REU) or a postbaccalaureate fellow (#RaMP)!
[thread!] Interested in protein design, deep learning, computational biophysics? Or computational RNA folding, vaccines, enzymes, antibodies, biomaterials, machine learning? Become a <a href="/RosettaCommons/">Rosetta Commons</a> undergraduate summer intern (<a href="/NSF/">U.S. National Science Foundation</a> #REU) or a postbaccalaureate fellow (#RaMP)!
[thread!]](https://pbs.twimg.com/media/FkC3MFjUcAACK8L.jpg)

We are hiring for a new Program Director for a Synthetic Biology Center that will focus on materials and engineering environmental microbes. The Center spans Stanford, MIT, Northwestern and several other institutions. Please email me or Christopher Voigt if it is of interest.

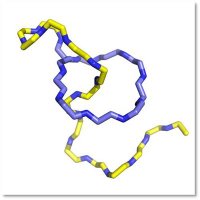
The latest Link lab paper is out J. Am. Chem. Soc. today led by Brian Choi: pubs.acs.org/doi/10.1021/ja…. In this work we show that ATP grasp enzymes that usually make esters can also make thioesters.








