
Jiangning (John) Song
@supercs08
Director of Data-driven Bioinformatics and Biomedicine Lab | Professor of Monash University | Assoc Editor of IEEE J Biomed Health Infomatics, BMC Bioinform
ID: 897040136109645824
https://research.monash.edu/en/persons/jiangning-song 14-08-2017 10:20:09
440 Tweet
364 Followers
701 Following

1/Unlocking the secrets of how the third form of life —archaea— makes energy, with potential applications for transitioning to a green economy. Published in Cell, led by scientists including Monash Biomedicine Discovery Institute's Chris Greening Jill Banfield Bob Leung More bit.ly/4c5KD1w


We want Monash University to be the place to be for microbiology in the post-metagenomics era. So much going on.



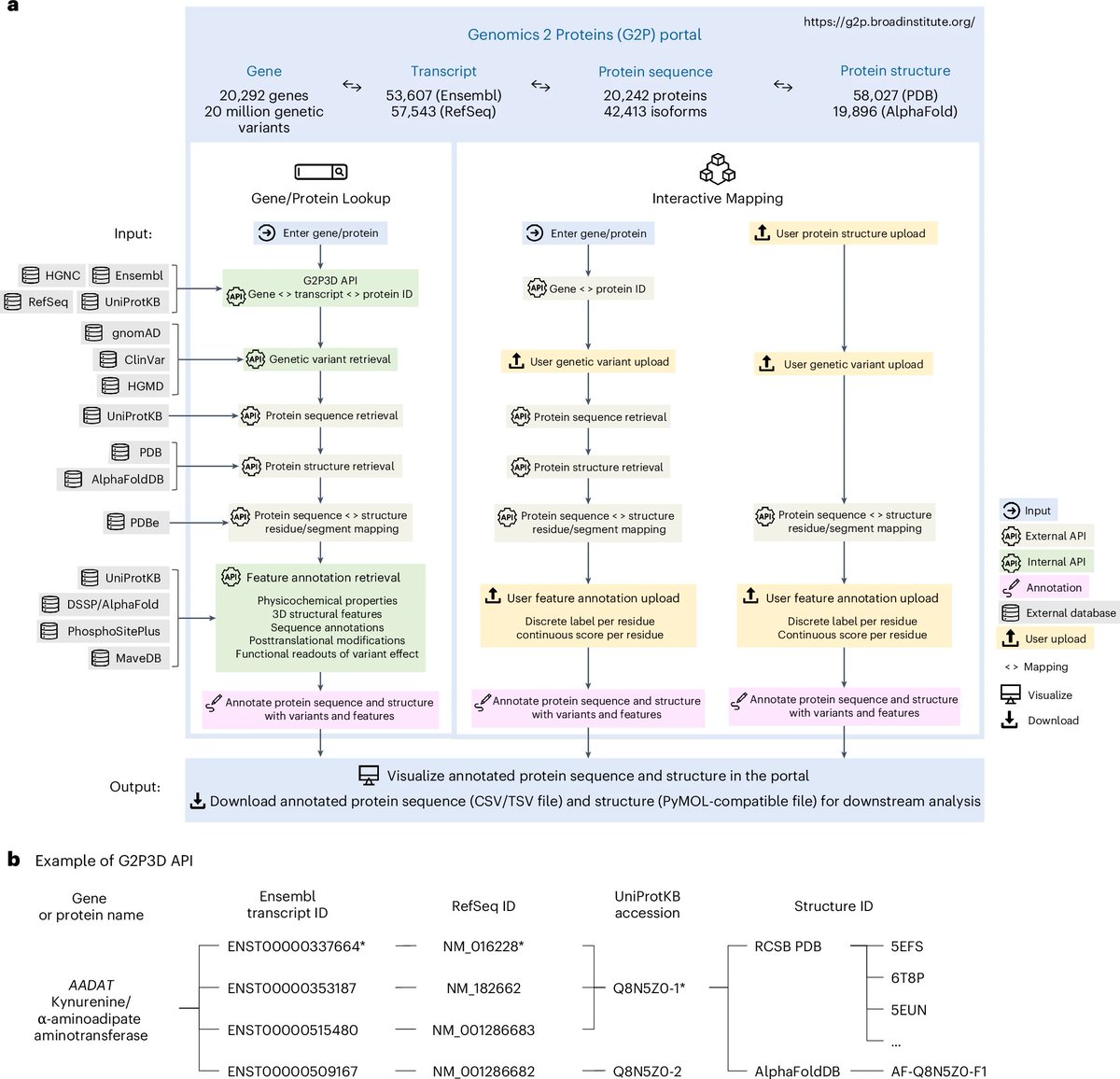
Genomics 2 Proteins portal: a resource and discovery tool for linking genetic screening outputs to protein sequences and structures | Nature Methods - Genomics 2 Proteins (G2P) portal, linking 20,076,998 genetic variants to 42,413 protein sequences and 77,923 structures (58,027






Happy to announce our special issue on "Application of large language models in genome analysis", now live on Genome Biology. Honored to serve as a guest editor alongside Jiangning (John) Song! We welcome your manuscript submissions to this groundbreaking collection.biomedcentral.com/collections/CO…




