Riggi Lab
@nriggi_lab
We are studying #Epigenetics at #Single Cell resolution and #3D Genome in #Cancer evolution/drug resistance @ Genentech.
ID: 803182301907734528
28-11-2016 10:22:37
164 Tweet
199 Followers
243 Following












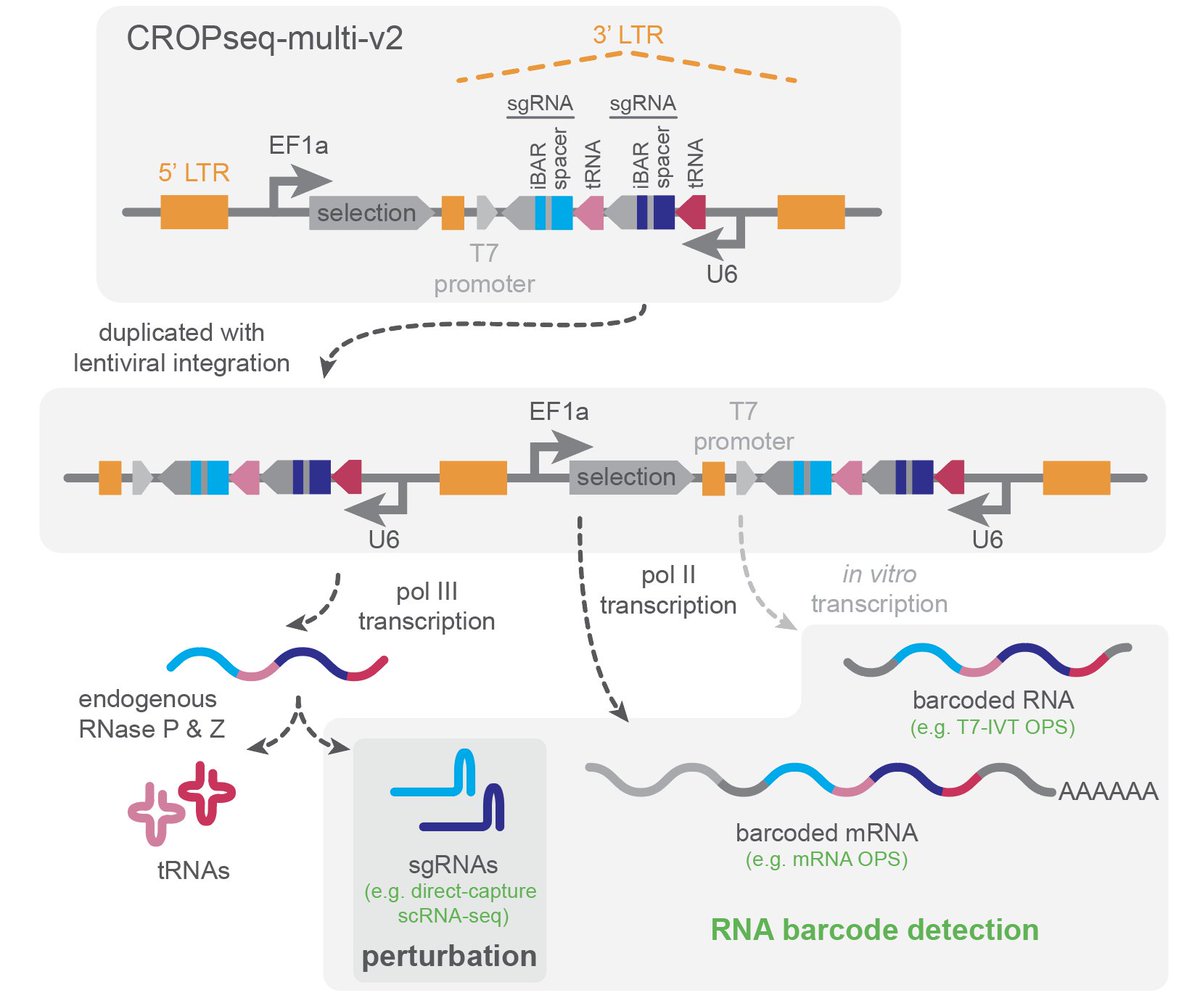
We’re Blainey Lab sharing a major update to CROPseq-multi, our versatile system for CRISPR screens that is compatible with individual and combinatorial perturbations, diverse SpCas9-based technologies, and multiple high-content, single-cell readouts. biorxiv.org/content/10.110…




Spatial-DMT is now published in nature ! For the first time, we can simultaneously map DNA methylation and RNA transcription in tissue context. Great collab with Wanding Zhou lab. Big congrats to Chin Chin , Hongxiang Hongxiang Fu and team! nature.com/articles/s4158…