Marcus D Wilson
@mdrwilson



We are delighted to establish our new Discovery Research Platform for Hidden Cell Biology The University of Edinburgh and grateful to Wellcome for support. Biological Sciences | University of Edinburgh @WTcell

Here is our latest work in which we explored the relationship between MeCP2, heterochromatin and DNA methylation using live-cell imaging. This is a collaboration with Tom Burdon The Roslin Institute, Thomas Jenuwein MPI of Immunobiology & Epigenetics and Claude Libert VIB. biorxiv.org/content/10.110…


EMPIAR.org/EMPIAR-11454 contains raw data for the "Structure of the Nucleosome Core Particle from Trypanosoma brucei", published @NAR_Open (doi.org/10.1093/nar/gk…) by the Marcus D Wilson lab The University of Edinburgh. #EMPIAR #cryoEM #NeglectedTropicalDiseases

We are very excited to share with you the results of great teamwork with @AmelieFT lab! In this study, two graduate students Edgar Pinedo-Carpio and @DessaptJcharacterized the critical role of FIRRM/C1orf112 in the disassembly of RAD51 during DNA repair. science.org/doi/full/10.11…

Job Alert📢📢 We are recruiting postdocs in Protein Biochemistry bit.ly/45vrx1A & Cell Biology bit.ly/45ijcOX to join our lab Department of Biochemistry at Oxford University. Join us to tackle the next big challenges in the Wnt signalling field in a fun, collaborative & supportive environment!



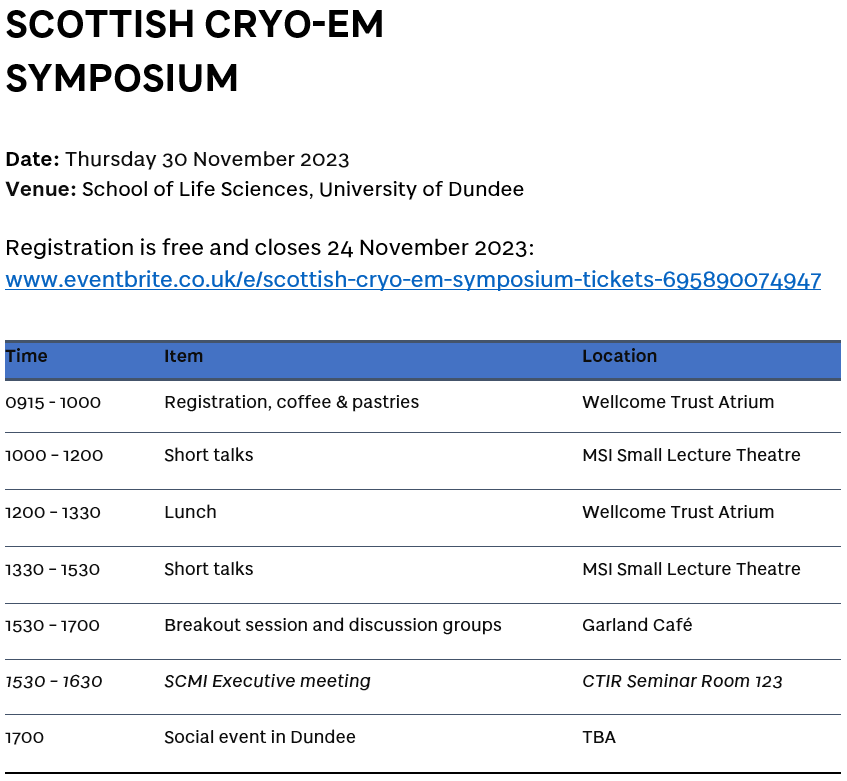
Join us for the first Scottish Symposium on Cryo-electron Microscopy !❄️🔬🏴eventbrite.co.uk/e/scottish-cry… SCMI UoD Life Sciences Centre for Targeted Protein Degradation SULSA co-organised with Mark A. Nakasone Tom Owen-Hughes Charlotte Crowe


Great talk by Hannah Wapenaar from Marcus D Wilson lab solving some beautiful structures and highlighting tips and tricks from data processing software packages for #cryoEM #Scottish_cryoEM23

The paper version of our DNMT3B study is now out in EMBO Reports 🥳: embopress.org/doi/full/10.10… You can read more on the previous thread but here is a summary of the main updates. This is one for Southern blot fans! 🧵1/9