
Eli Mann
@manntis4
ML for Chemistry at @RowanSci
ID: 1790070747836121088
13-05-2024 17:26:13
24 Tweet
49 Followers
89 Following


The first paper citing Egret-1 was released recently, less than a month after we released our model... Sanketh Vedula, Alex Bronstein, + co-workers use Egret-1 embeddings to predict local protein properties like NMR shift + secondary structure.
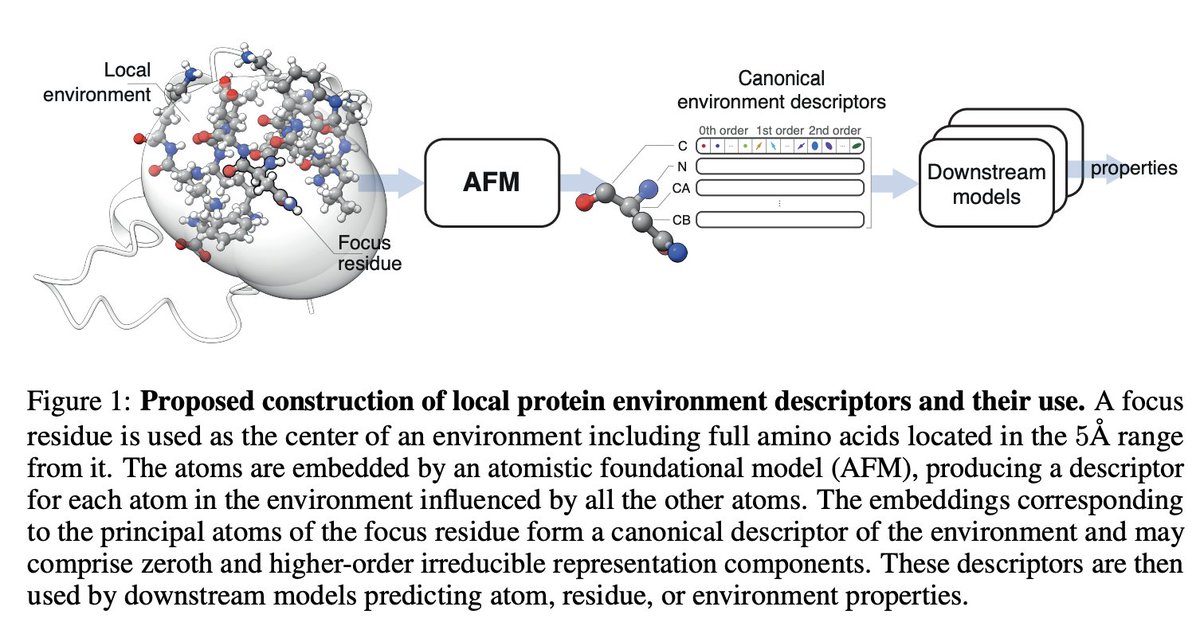


Really cool work by Meital and Sanketh Vedula, excited to highlight this. Check out their blog post and paper here!


The long-anticipated g-xTB paper was just released on ChemRxiv. g-xTB is the next semiempirical method from Grimme and co-workers at Rheinische Friedrich-Wilhelms-Universität Bonn. (I've heard rumors about this work for almost two years.) Here's some quick thoughts upon an initial read:


Our new ChemRxiv preprint: AIMNet2-rxn: A #MachineLearning Potential for Generalized Reaction Modeling on a Millions-of-Pathways Scale! #compchem Collaboration with Brett Savoie group. Funded by Office of Naval Research (ONR) under #MURI program chemrxiv.org/engage/chemrxi…



Chai Discovery (Joshua Meier Chai Discovery) just released Chai-2, a model for "zero-shot antibody design in a 24-well plate." The most existing result? Chai-2 successfully generated binders for 50% of the 52 targets they assayed (100x better than SOTA)! Some thoughts below:





hydrogen-bond-donor strength workflow now available on rowan! nice work Corin Wagen

Great to see Rowan's redox potential and Fukui-index calculations used to help with total synthesis. Congrats Matt Carson Kozlowski Lab and co-workers!







