
Mingliang Jin
@jin_mingliang
cryoEM postdoc UCSF
ID: 1416987916220260355
19-07-2021 05:07:09
293 Tweet
140 Followers
543 Following





DiffModeler: large macromolecular structure modeling for cryo-EM maps using a diffusion model Nature Methods • DiffModeler is an automated tool that excels at modeling large protein complexes from cryo-EM maps, particularly at intermediate resolutions (5-10 Å). It offers a



Pleased to introduce DiffModeler, a new tool for str modeling for cryo-EM appeared in Nature Methods this week! Uses diffusion model to emphasize main-chains in maps and fits Alphafold models. Works up to 10-15 A res. nature.com/articles/s4159… Server: em.kiharalab.org


🍂NEW VIDEO🍂 All through Autumn, trees race against time to recycle their chlorophyll--and basically everything else in their leaves. But why go through all this effort? The answers are honestly mind-blowing: rendered in #blender3d using Brady Johnston's #molecularnodes


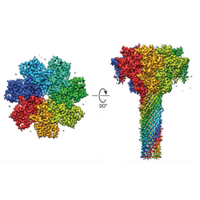




A novel cryo-EM affinity-grid approach utilizing monodispersed single-particle streptavidin on a lipid monolayer is introduced to improve particle absorption, thereby addressing challenges like preferential angular distribution rdcu.be/d1MNM eBIC Beamline STRUBIOxford





