Mina Shaigan - @minashaigan.bsky.social
@minashaigan
Ph.D. student at the Institute of Computational Genomics @vanvanka123, University Hospital @RWTH Aachen
#timeSeries #epigenomics #probabilisticMachineLearning
ID: 1494038531160821762
http://minashaigan.github.io 16-02-2022 19:58:51
87 Tweet
44 Takipçi
158 Takip Edilen



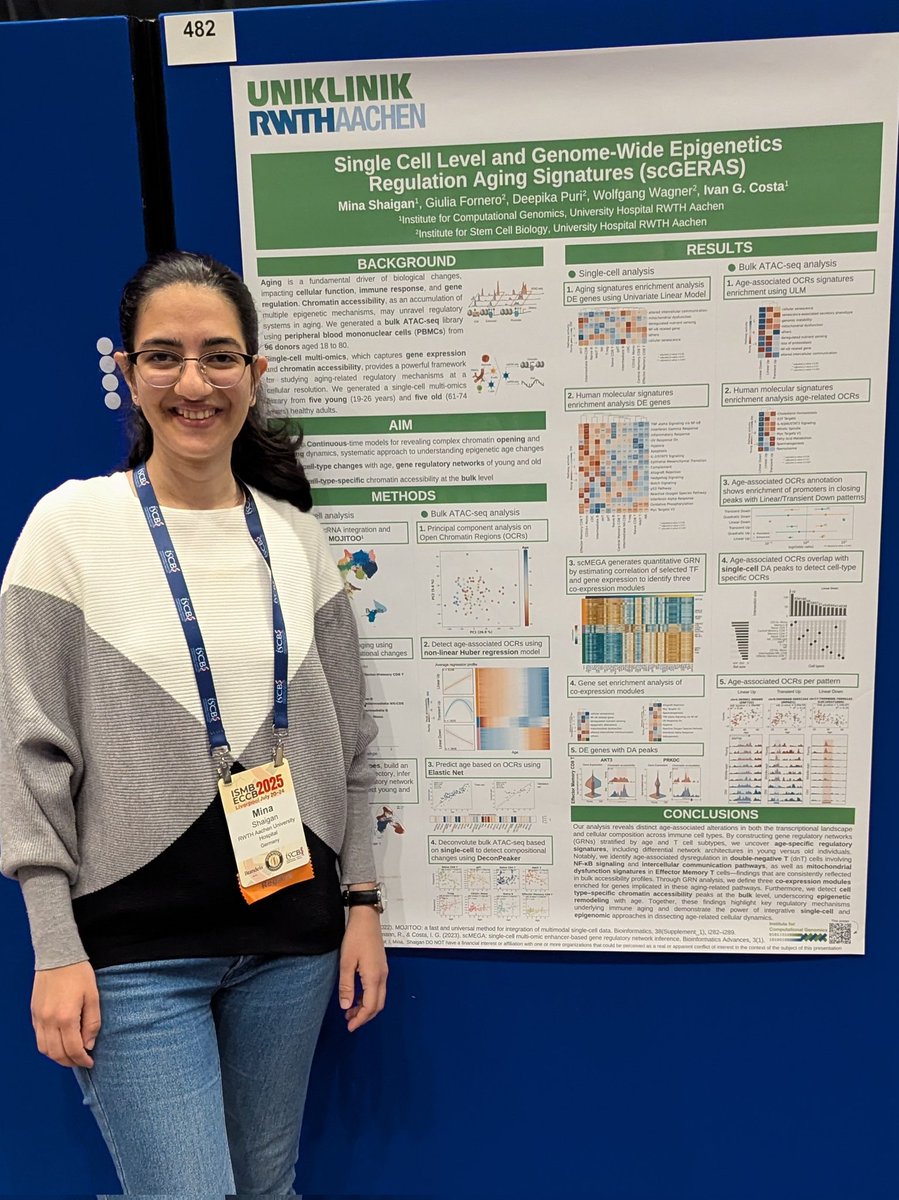
I'm so delighted that I shared our work with Mehdijoodaki and Costa Lab - @costalab.bsky.social at ISCB News #RSGDREAM 2024 in Madison this October on the disease trajectory detection using PILOT check out our paper embopress.org/doi/full/10.10… #SystemsBiology #singlecell #pathomics


Thrilled to share PHLOWER, which uses Hodge Laplacians to build trajectory-level embedding for inferring complex cellular differentiation trees from multimodal (RNA+ATAC) single-cell data by Mingbo Jitske Jansen with Rafael Kramann, MD PhD Christoph Kuppe tinyurl.com/yvu48j6k 1/7

Polars is between 10 and 100 times as fast as pandas for common operations and is actually one of the fastest DataFrame libraries overall. blog.jetbrains.com/pycharm/2024/0… via PyCharm, a JetBrains IDE?

Really nice to contribute to this study by Kleger Lab with Mina Shaigan - @minashaigan.bsky.social showing that a composite mutation (gene truncation in one locus and a enhancer deletion in the other locus) of the transcription factor Onecut1 causes early onset diabetes. cell.com/cell-reports/f…
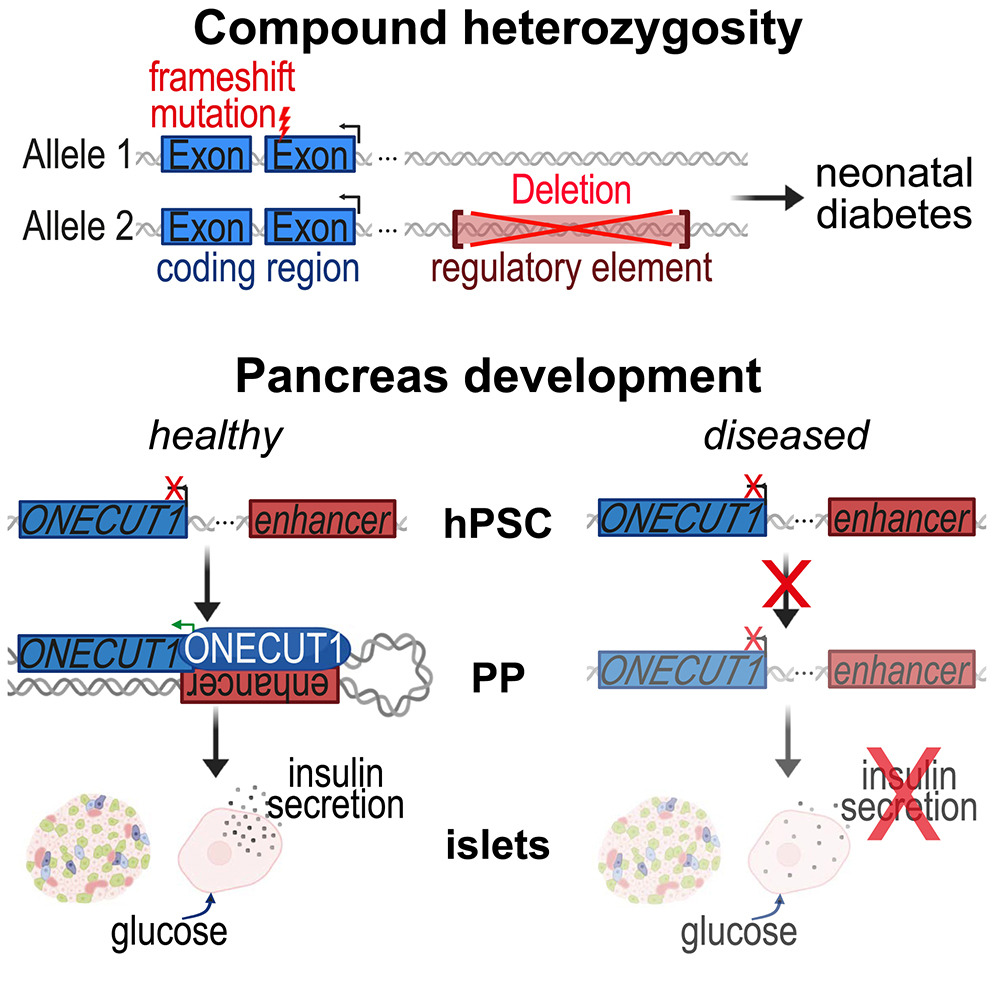

Kleger Lab Mina Shaigan - @minashaigan.bsky.social This also builds up on previous work where we observed several GWAS hits affecting TF binding sites in enhancers of ONECUT1 nature.com/articles/s4159…







📢 Attending #ISMBECCB2025 in #Liverpool ? Don’t miss our #tutorial:"Computational approaches for cell-cell communication from #singlecell & #spatialtranscriptomics" in collaboration with CostaLab Costa Lab - @costalab.bsky.social 🗓 July 15 | 🕘 09:00 BST | 💻 Virtual 🔗 iscb.org/ismbeccb2025/r…










![Mathieu (@miniapeur) on Twitter photo [1/5] Data often has inherent geometric and topological properties that can be exploited by machine learning algorithms. Below are four areas of machine learning that explore this important aspect. [1/5] Data often has inherent geometric and topological properties that can be exploited by machine learning algorithms. Below are four areas of machine learning that explore this important aspect.](https://pbs.twimg.com/media/Gu3un7gXwAAUX5o.jpg)