
Carlos Mora
@carlosm49544824
Biologist
ID: 1263502619508703232
21-05-2020 16:11:24
1,1K Tweet
85 Takipçi
730 Takip Edilen

Why are evolved networks (such as gene and metabolic webs) different from engineering designs? One lesson is provided by generating electronic circuits using Darwinian evolution, as shown in this Scientific American paper by John Koza Víctor de Lorenzo Michael Levin jstor.org/stable/pdf/260…



Great, we are now also in Science! GeneticsUB Universitat de Barcelona Facultat de Biologia UB science.org/content/articl…
PhyloAln: a convenient reference-based tool to align sequences and high-throughput reads for phylogeny and evolution in the omic era academic.oup.com/mbe/advance-ar… Molecular Biology and Evolution

Nice - cohesin may only be required for enhancer-promoter comm for distal enhancers (>50kb). This nicely extends (and generalises) findings from Wendy Bickmore at shh locus, tethering a TF at different distances. There 100kb was the tested cut off - interesting to see even 50kb


FoldMason progressively aligns thousands of protein structures in seconds, enabling remote MSA for distant phylogeny. Highlights: structural flexible MSA, LDDT conservation score, friendly webserver🧵 💾github.com/steineggerlab/… 🌐search.foldseek.com/foldmason 📄biorxiv.org/content/10.110…



Our latest: "Reverse development in the ctenophore Mnemiopsis leidyi" out in bioRxiv, Pawel Burkhardt Michael Sars Centre Universitetet i Bergen. Check it out: biorxiv.org/content/10.110… Illustrations by talented Nicholas Bezio (Bezio Studios) . [🧵1/6]
![Joan J. Soto-Angel (@joanjsoto) on Twitter photo Our latest: "Reverse development in the ctenophore Mnemiopsis leidyi" out in <a href="/biorxivpreprint/">bioRxiv</a>, <a href="/Pawel_Burkhardt/">Pawel Burkhardt</a> <a href="/MSarsCentre/">Michael Sars Centre</a> <a href="/UiB/">Universitetet i Bergen</a>. Check it out: biorxiv.org/content/10.110…
Illustrations by talented <a href="/NickBezioArt/">Nicholas Bezio (Bezio Studios)</a> . [🧵1/6] Our latest: "Reverse development in the ctenophore Mnemiopsis leidyi" out in <a href="/biorxivpreprint/">bioRxiv</a>, <a href="/Pawel_Burkhardt/">Pawel Burkhardt</a> <a href="/MSarsCentre/">Michael Sars Centre</a> <a href="/UiB/">Universitetet i Bergen</a>. Check it out: biorxiv.org/content/10.110…
Illustrations by talented <a href="/NickBezioArt/">Nicholas Bezio (Bezio Studios)</a> . [🧵1/6]](https://pbs.twimg.com/media/GU4laHtXIAA9kcb.jpg)



There's a new paper by Simone Tiberi et al. from the Mark Robinson @[email protected] lab on a method for finding differentially regulated genes: "spliced and unspliced (or precursor) mRNA..can be used to study gene regulation and changes in gene expression production" 1/ nature.com/articles/s4159…

Major effort from the lab (💪Robert Frömel), finally on biorxiv: We built 60,000 fully synthetic enhancers and measured their cell state specific activity during blood stem cell differentiation. Why did we do that and what did we learn🧵? biorxiv.org/content/10.110…


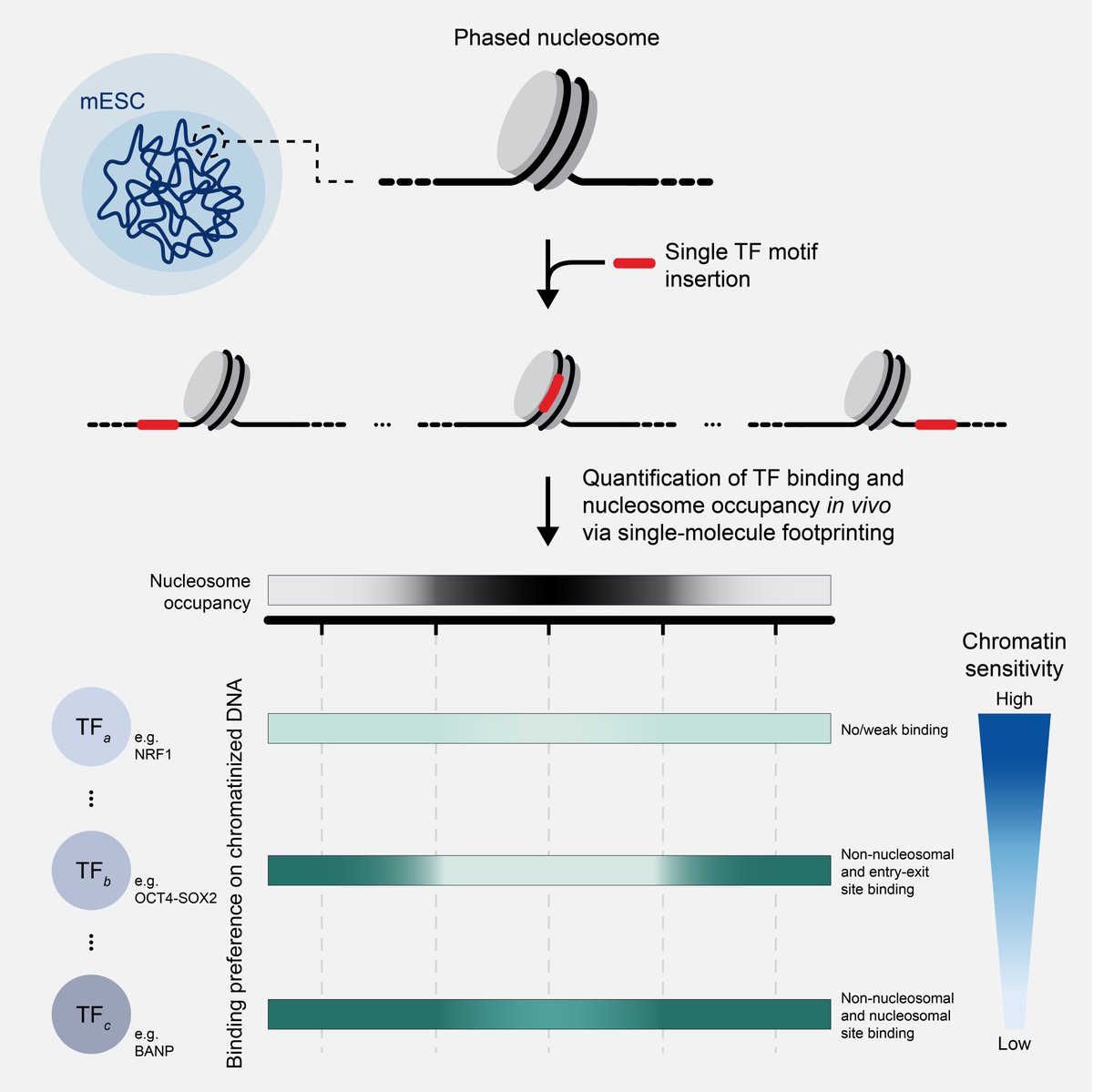
Wouldn’t it be great to test how individual transcription factors engage with their motif in the nucleus? We tried exactly that in a study lead by Ralph (Ralph Grand) and Marco (Marco Pregnolato) now out in Molecular Cell authors.elsevier.com/sd/article/S10…. 1/7

Happy to share our new paper on the characterization of MSTV1, an archaeal virus infecting Methanobrevibacter smithii, the dominant archaeon in the human gut. This was a beautiful collaboration between Mart Krupovic and Simonetta Gribaldo labs. 👇🧵nature.com/articles/s4146…

I am thrilled to announce I have secured an European Research Council (ERC) Starting Grant (1.5 MILLION euros!) to study the #genomics of miniaturisation in vertebrates! Press release: news.ku.dk/all_news/2024/…






