Heiko Runz
@heikorunz
genetics, medicine, drug discovery, ethics
ID: 2774101913
18-09-2014 01:46:04
437 Tweet
466 Followers
15 Following

A rare protein coding variant study identifies six significant hand grip strength genes, KDM5B, OBSCN, GIGYF1, TTN, RB1CC1, and EIF3J. Nature Communications nature.com/articles/s4146…

Our new paper is now in biorxiv: We modified the Illumina library prep & introduce RIMS-seq2 which enables DNA sequencing and regional methylation calls in humans through a single, easy experiment. New England Biolabs, Albert Vilella Phase Genomics (1/4) biorxiv.org/content/10.110…


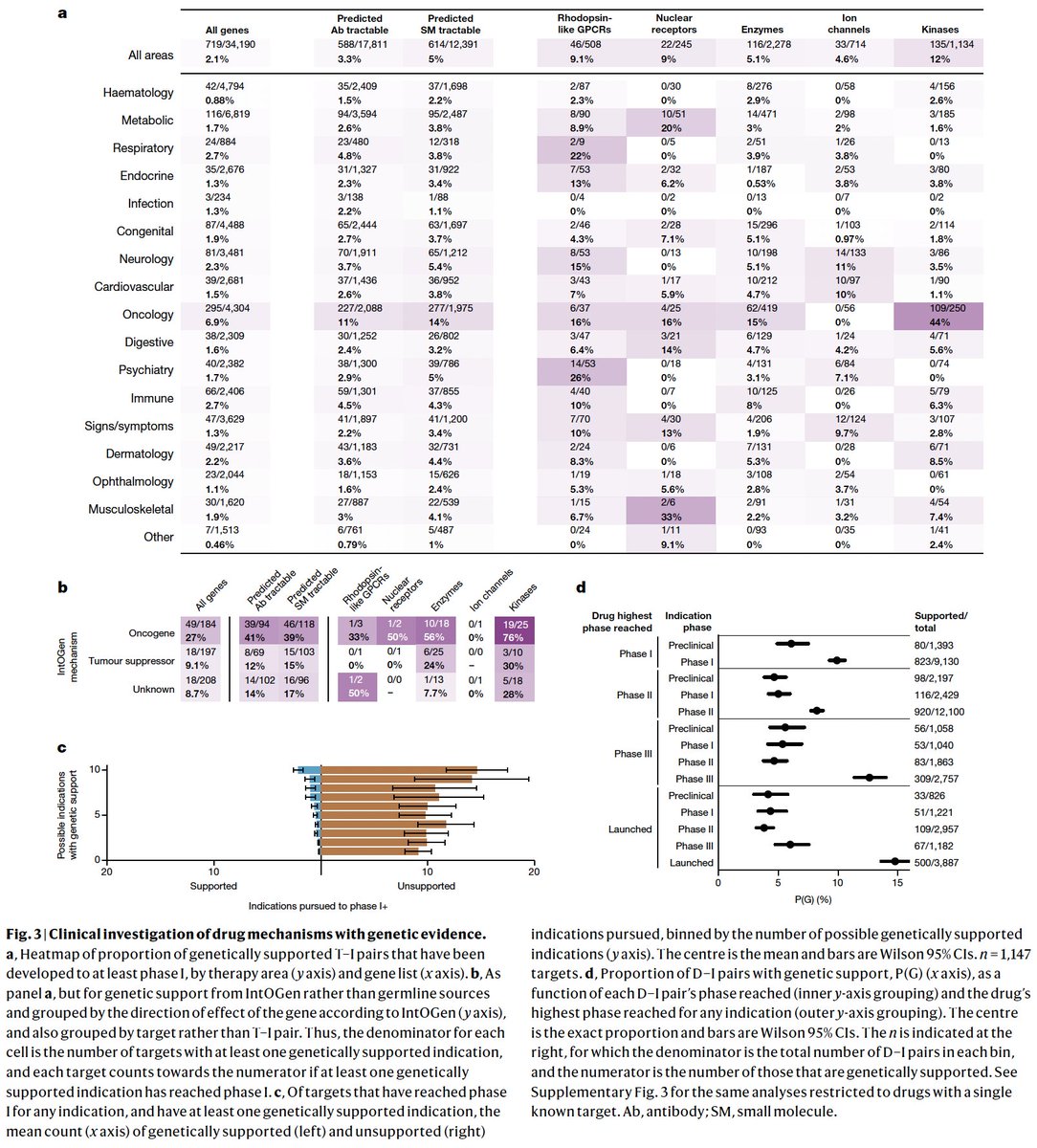
Is having even more gene-disease associations still useful for picking drug targets? For which types of programs does it matter? Has pharma shifted focus towards genetically validated targets? New paper by me, Matt Nelson, Coco (Chengliang) Dong, & Jeffery Painter doi.org/10.1101/2023.0…👇


isGWAS: a new tool for ultra-fast association testing from just summary statistics ⬇️compute limitations: “green GWAS” for everyone & predicts “future” GWAS loci 👏to a fantastic collaboration: Optima MarioniGroup Benjamin Sun Please test, share thoughts: optima-isgwas.com

Our guide how to realize clinical recall studies from the amazing FinnGen resource (exemplified by a small pilot for Alzheimer's disease) has now been published in Scientific Reports. Thank you to all co-authors and participants! Valtteri Julkunen Eero Vuoksimaa nature.com/articles/s4159…




Super excited a series of THREE UKBIOBANK UKBIOBANK #proteomics papers are now online on nature! nature.com/articles/s4158… - flagship paper and two sister companion pieces: nature.com/articles/s4158… and nature.com/articles/s4158… - Herculean team effort across so many teams! More to follow.

Thrilled to see our paper on rare variant associations with plasma protein levels in the UK Biobank published in @nature today nature.com/articles/s4158… Summary statistics are publicly available at azphewas.com and an interactive portal astrazeneca-cgr-publications.github.io/pqtl-browser/i…




Exciting news! Our latest study has been published in Nature Communications: nature.com/articles/s4146… Congrats and huge thanks to Chia-Yen Chen, Heiko Runz, and all our other co-authors, Tian Ge, Todd Lencz, Max Lam, Jimmy Liu, Ellen Tsai for their exceptional contributions!




Excited to share our work now in Nature Aging. Multiplex blood #proteomics + #MachineLearning in UK Biobank improve multiple #disease incidence #prediction beyond traditional risk factors. nature.com/articles/s4358… Shout out to my colleagues and old team at Biogen

Delighted to see Robert Hillary's protein vQTL + GxE paper using data from UK Biobank published in Nature Communications. nature.com/articles/s4146…


Excited to share our work now in Nature Communications. Variance protein QTLs in UK Biobank uncover gene interactions spanning female #physiology, #blood groups, white cell indices. nature.com/articles/s4146… Shout out to my co-authors and former Biogen team.
![Sasha Gusev (@sashagusevposts) on Twitter photo [Chen et al. 2023 NG; pubmed.ncbi.nlm.nih.gov/37231097/] looked for rare coding associations with three phenotypes: Edu, Reaction Time, VNR. They found 5 associations with Edu (+3 others), demonstrating that Edu can be a reasonable and accessible proxy phenotype for rare var studies. [Chen et al. 2023 NG; pubmed.ncbi.nlm.nih.gov/37231097/] looked for rare coding associations with three phenotypes: Edu, Reaction Time, VNR. They found 5 associations with Edu (+3 others), demonstrating that Edu can be a reasonable and accessible proxy phenotype for rare var studies.](https://pbs.twimg.com/media/F6a1ra8XsAAF3rE.jpg)