
François Aguet
@francoisaguet
Computational Biologist | Head of computational biology @PredictaBiosci | Previously group leader @illumina AI lab and @broadinstitute
ID: 1162143901
http://francoisaguet.net 09-02-2013 06:17:32
56 Tweet
381 Takipçi
162 Takip Edilen

The cell type work in GTEx was led by Sarah Kim-Hellmuth who, together with François Aguet, computationally estimated the enrichment of 64 cell types in all GTEx samples & mapped cell type interacting eQTLs and sQTLs for 7 cell types. science.sciencemag.org/lookup/doi/10.… science.sciencemag.org/content/369/65…

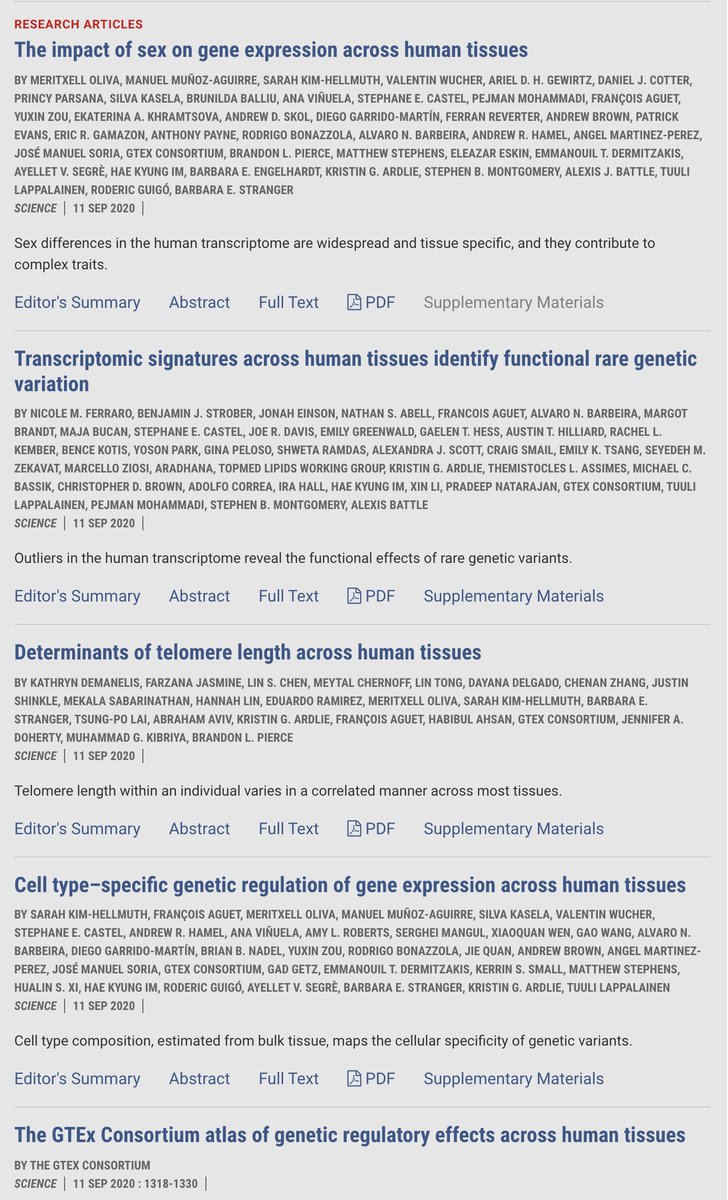
Hugely excited that our GTEx paper on cell type specific genetic regulation of gene expression across human tissues is finally out in Science Magazine : science.sciencemag.org/content/369/65…

By taking a deeper dive into GTEx Portal data, François Aguet, Sarah Kim-Hellmuth et al show how much cell type matters when thinking about variants' effects on gene expression. broad.io/GTEx #GTEx

The final set of analyses from the GTEx Consortium have just been published: bit.ly/gtexScience broad.io/BC09112020GTEx. All of the GTEx V8 public data is available GTEx Portal and protected data on anvil.terra.bio #eqtl

A banner day for human #genomics: advancing the understanding of the activity and regulation of our ~20,000 protein-coding genes ★ Extraordinary work by Tuuli Lappalainen Manolis Dermitzakis François Aguet &🌎colleagues 🙏 Summary sciencemag.org/news/2020/09/i… Science Magazine Science Visuals

Why do >10% of older adults have clonal blood mutations? Simultaneous germline & somatic genome analysis in ~97K pinpoints causal mechanisms. This work w/ Pradeep Natarajan, Sek Kathiresan MD, @J__Stock now online @nature: nature.com/articles/s4158… Tag-team tweetorial w/Pradeep Natarajan! 1/n


We are close to launching a new feature where GTEx Portal v8 protected data can be downloaded without egress fees (you still need dbGaP approval). We are looking for beta testers - DM me for details. National Human Genome Research Institute NIH Common Fund Tuuli Lappalainen Alexis Battle François Aguet #ASHG2020

We are excited to announce you can now download the raw GTEx protected data from AnVIL without egress fees! You will still need dbGaP approval but this will streamline novel data integration and analysis for all GTEx Portal National Human Genome Research Institute NIH Common Fund anvilproject.org/news/2020/11/2…

WE ARE HIRING! Come join us in the Getz Lab @BroadInstitute/Mass General Cancer Center to help shape the future of #CancerResearch at the cutting edge of #ComputationalGenomics and cancer biology! Links in thread👇👇👇 (1/5)


We are very excited to share our single-nucleus cross-tissue atlas w/ Eugene Drokhlyansky, François Aguet, Shankara Anand, Ayshwarya Subramanian, Evgenij Fiskin, Ayellet Segrè, Orit Rozenblatt-Rosen, Kristin Ardlie, Aviv Regev and many others 1/n



Our single-nucleus cross-tissue cell atlas is out in Science Magazine as part of Human Cell Atlas: science.org/doi/10.1126/sc… w/ Eugene Drokhlyansky, Shankara Anand, Evgenij Fiskin, Ayshwarya Subramanian, François Aguet, Ayellet Segrè, Orit Rozenblatt-Rosen, Kristin Ardlie, Aviv Regev & many others 1/


Excited to share our paper, out today in nature, where we characterize transcriptome variation in human tissues by long-read sequencing. Huge thanks to Dafni Beryl Cummings Garrett Garborcauskas Daniel MacArthur and other collaborators. 🧵 nature.com/articles/s4158…

Super excited to share our paper published today in nature, where we discover the primary role of ADAR RNA editing in inflammatory diseases. Huge thanks to Mike Mike Gloudemans, Jonathan Jonathan Pritchard, Stephen @sbmontgom, Billy Billy Li and other collaborators. nature.com/articles/s4158…

Hot off the press! Check out our new ms in collaboration with Dr. Catherine Wu’s lab at Dana-Farber #DFCI and colleagues from Spain, Germany and the US on creating a molecular map of #CLL from #multiomics data of >1100 patients that improves prognostics! nature.com/articles/s4158…
