
Moritz Ertelt
@erteltmoritz
Postdoc @FraunhoferITMP
Biologist designing proteins
ID: 1227799029666287616
13-02-2020 03:38:14
86 Tweet
228 Followers
641 Following


Our work validating designed membrane b-barrels in silico is published! It started as Elixir #3DBioinfo meeting discussion (I generated models instead of preparing my talk), then a fun exchange with Sergey Ovchinnikov. Thanks to all! VIB_StructuralBiologyBrussels ELIXIR Belgium onlinelibrary.wiley.com/doi/10.1002/pr…

Today in nature we present the results of our exploration of the soluble protein universe and the design of functional soluble membrane proteins with Casper Goverde Nico Goldbach Bruno Correia 1/6 nature.com/articles/s4158…

Yesterday, I had the chance to present my first spotlight poster at #MoML2024 at Mila - Institut québécois d'IA AI Institute! I presented joint work done with Luisa Kärmer and our amazing advisors, Julia Westermayr and Jens Meiler Lab. Huge thanks to @Esporascicomm for the amazing graphic design work!




Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates. Bruno Correia Sergey Ovchinnikov biorxiv.org/content/10.110…


Antibodies are highly diverse, but most possible sequences are unstable or polyreactive. In this work led by Milind Jagota, we propose a new source of data for modeling constraints from these properties. Our models show clear improvements in predicting antibody dysfunction.(1/n)
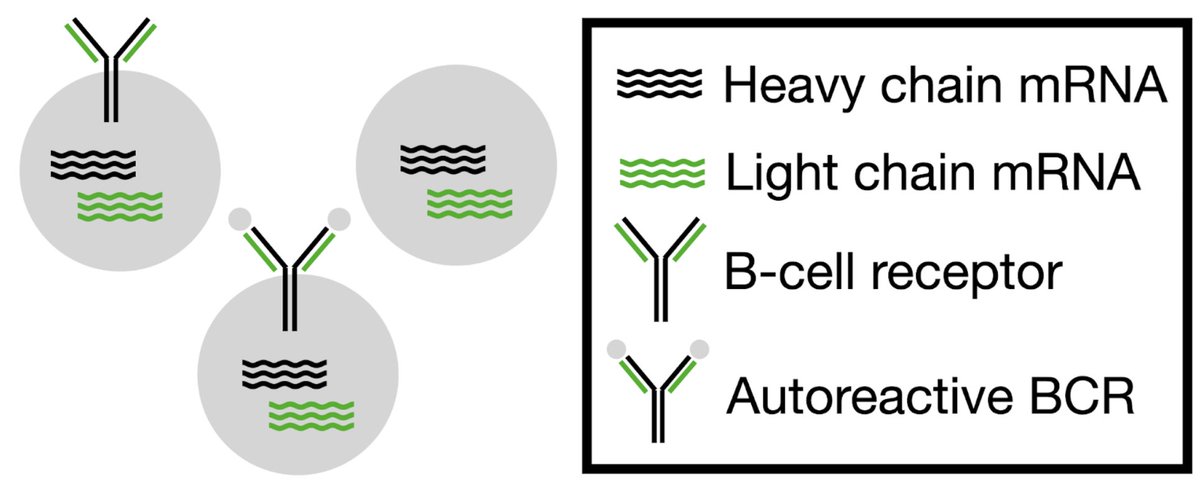







I’m happy to announce my latest paper has been released as a preprint on BioRxiv: Predictions from Deep Learning Propose Substantial Protein-Carbohydrate Interplay. This paper was only able to happen thanks to both Jeffrey J. Gray and Ronald Schnaar doi.org/10.1101/2025.0…








