Yang I Li
@yang_i_li
Human genomics, gene regulation, RNA. Associate Professor @UChicago.
ID: 1071972253
https://thelilab.com/ 08-01-2013 20:50:50
613 Tweet
1,1K Followers
592 Following




Yang I Li @BenjaminFair3 Carlos Buen Abad very interesting. Our 10y effort (though very low throughput) of characterizing many isoform-level functions seems to agree with that. isoforms differ in stability either at RNA or PROTEIN levels to affect total protein outputs 👇sciencedirect.com/science/articl… nature.com/articles/s4146…


"stay curious and excited about science, especially about what’s happening outside your field. You never know where your next good idea will come from." -Judy Lieberman Journal of Experimental Medicine doi.org/10.1084/jem.20…


Really impressive paper from Satoshi Koyama, Pradeep Natarajan, and co. looking at rare exome variants in 1.15M (MVP, UKB, AoU) individuals for blood lipids. Fig. 2a provides pretty strong support for including cryptic splice variants (from Illumina's SpliceAI) in gene-based tests.



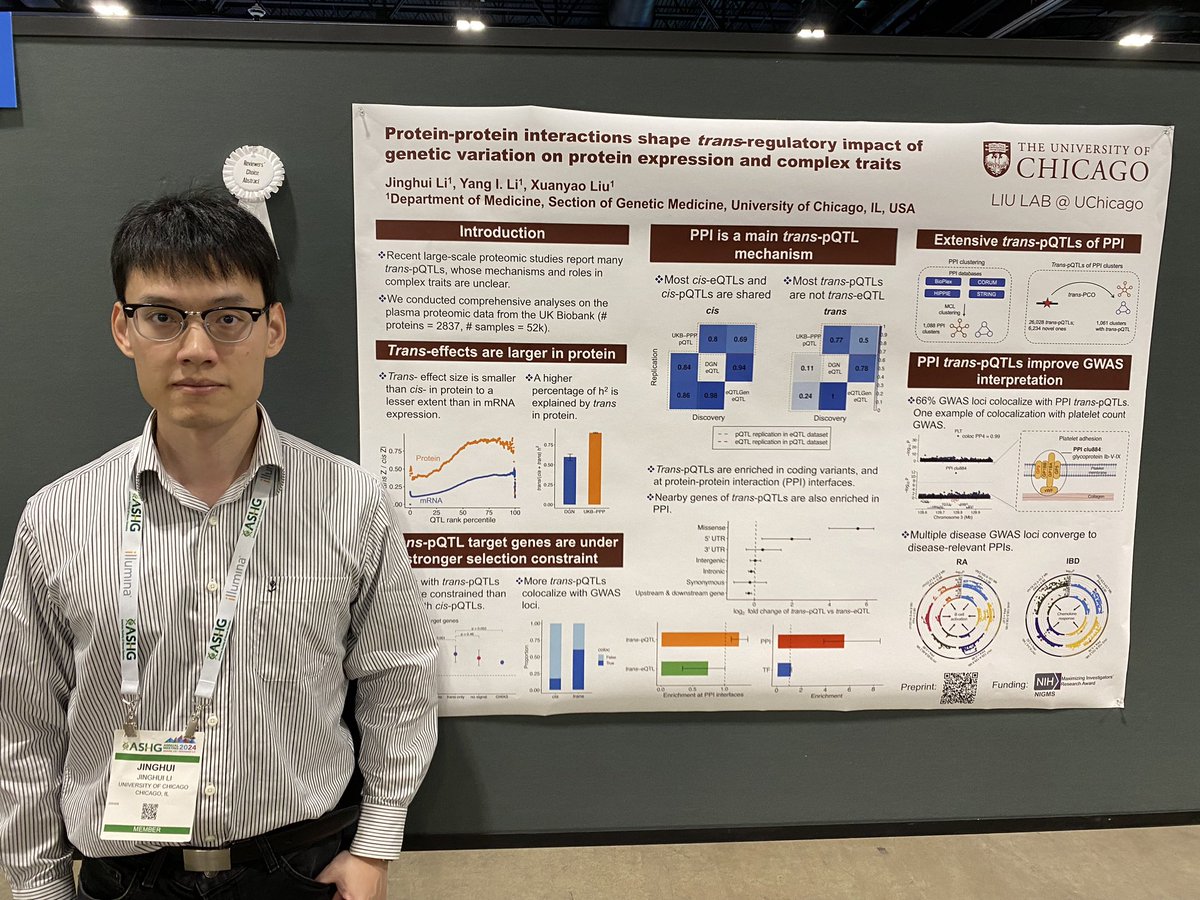
🚨New Preprint Alert: Our work led by Jinghui Li is up on bioRxiv. We re-analyzed large maps of trans-pQTLs, and found that protein–protein interactions shape trans-regulatory impact of genetic variation on protein expression and complex traits. A brief thread🧵👇





Our work is out in AJHG! I wholeheartedly thank all the co-authors marie.saitou Andy Dahl Qingbo Seiha Wang for their time and effort! All four of us started our own labs between 2019 and 2021 and the pandemic didn't make things easy. Super proud of everyone.

Super thrilled to see Zepeng (Phoenix) Mu's work out in preprint form! See thread for summary of his thorough mapping and analysis of caQTLs from PBMC scATAC-seq data.

My Chinese American colleagues in Texas public universities such as the whole U Texas including MD Anderson are now required to report family visit trip to China ahead of trip, and provide a post-travel briefing detailing the trip. Land of the free (your experience may differ).