Silvana Konermann
@skonermann
Cofounder @arcinstitute and Assistant Professor @Stanford. HHMI Hanna Gray Fellow. CRISPR, RNA and Alzheimer’s via @salkinstitute, @MIT and @eth
ID: 913611794
30-10-2012 01:13:47
234 Tweet
8,8K Followers
538 Following





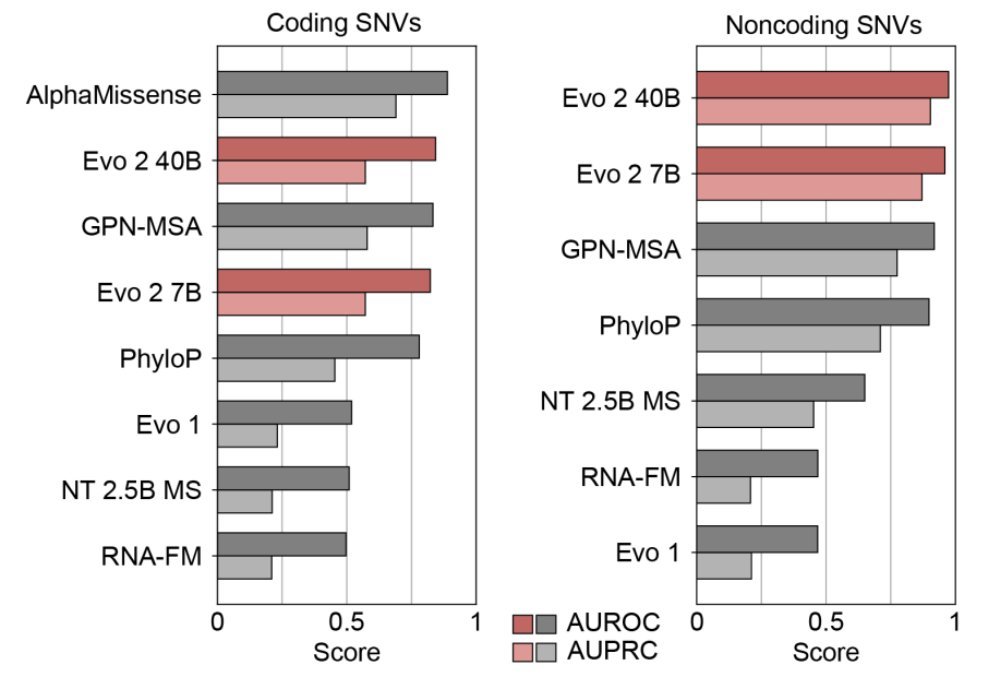
New from Arc Institute: Evo 2, the largest (by training compute) biology ML model ever, and one of the largest-ever open source ML models in any category. Evo 2 is a foundation model trained on 9T DNA base pairs that learns a lot of fundamental details about life. A few examples



We made an AI agent go vroom on all of SRA to create scBaseCamp, Arc Institute's ever-expanding and uniformly processed single cell data repo! 230M cells drawn from 21 species, 72 tissues, and counting... arcinstitute.org/manuscripts/sc… x.com/yusufroohani/s…

Spurred by how well things worked with Evo 2, and with the awesome people who helped out, I'm curious if there's interest in a larger Arc Institute software engineering volunteer program. Something like: • Spend 6–12 months working full-time at Arc. • Learn/perform

Joining forces with Babak Alipanahi, Hormozdiari Lab, Aiden M Sababi & Mehran Karimzadeh to bring you Exai-1; a multi-modal cell-free RNA foundation model for blood surveillance and liquid biopsy. Kudos to the broader exai bio team for getting this across the line. biorxiv.org/content/10.110…