Matteo Ferla
@matteoferla
Computational (bio)chemist in OPIG & XChem, University of Oxford
blog: blog.matteoferla.com
Michelanglo: michelanglo.sgc.ox.ac.uk
Opinions my own.
ID: 2973245047
http://www.matteoferla.com 10-01-2015 23:01:19
277 Tweet
554 Followers
504 Following


thanks to everyone who helped with the study which includes GECIP team GeneDx Prof Amaka C Offiah Melita Irving Reza Maroofian Baylor Genetics NIHR Oxford Biomedical Research Centre and Matteo Ferla (sorry your lovely fig had to move to suppl)


At the NIHR Oxford Biomedical Research Centre open day. New t-shirts, new venue, same strawberry 🍓 dna demo




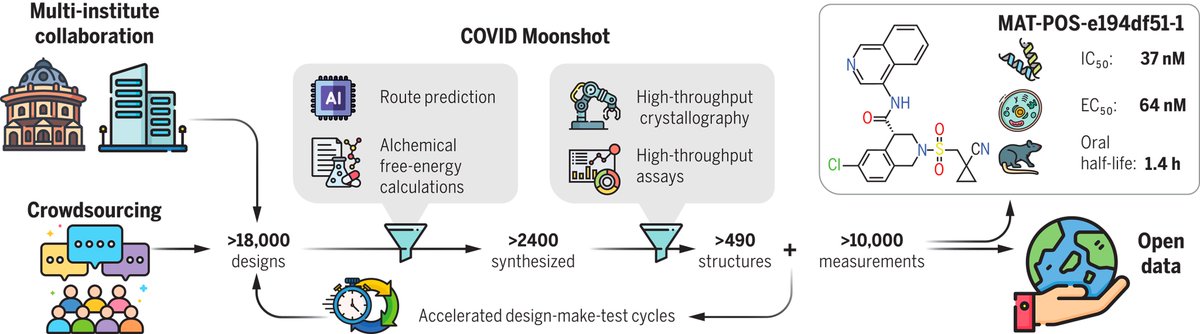
New & impressive Science Magazine Here's how you discover a potent new drug vs Covid 212 scientists/15 countries Open-science, patent free, high-throughput structural biology based design, #AI COVID Moonshot Melissa Boby Frank von Delft Daren Fearon science.org/doi/10.1126/sc…


The COVID Moonshot paper is out in Science: but can it help you as a compchem algo testset? Yes! All steps (frag to lead) crystallised & assayed, incl. negatives. Diverse methods, 4 series. Don't take my word, have a gander yourself: fragalysis.diamond.ac.uk/viewer/react/p…



Before the break I presented at the AViDD forum (asapdiscovery.org/forum/) for which I am very grateful for, but totally forgot to share the link: youtube.com/watch?v=kieDWY… (Title: Advancing Fragment-Based Drug Discovery in the ASAP Discovery consortium with Fragmenstein)






OPIG DPhil student Gemma Gordon led work to build and analyse "PLAbDab-nano: a database of camelid and shark nanobodies from patents and the literature". Just released on bioRxiv and available as an OPIG webapp: Preprint: biorxiv.org/content/10.110… Webapp: opig.stats.ox.ac.uk/webapps/plabda…

DPhil student Ollie Turnbull has built pIg-Gen, a language model for paired-chain antibody generation. It creates diverse, natural-like distributions of antibodies w/ similar biophysical properties to therapeutics. bioRxiv: biorxiv.org/content/10.110… Code: github.com/oxpig/p-IgGen


OPIGlets are out in force at #AIChem24, organised by our very own Garrett M. Morris DPhil students Lucy Vost, Arun Raja and Ísak Valsson are presenting posters, and Yael is giving a talk about her recent work MolSnapper on Wednesday! Please come and chat to them!