Junwei Shi
@junwei_s
Associate Professor at UPenn
ID: 1263159325
https://shipennlab.wixsite.com/shi-lab 12-03-2013 23:54:04
390 Tweet
1,1K Followers
507 Following


Penn Epigenetics UPenn CDB PennSysPharmATT Univ. of Pennsylvania Department of Cancer Biology Children's Hospital UPenn Dept of Biochem & Biophys Penn Medicine E. John Wherry Berger Lab Junwei Shi Improved gene editing method could power the next generation of cell and gene therapies #NBTintheNews via Penn Medicine pennmedicine.org/news/news-rele…

Penn Epigenetics UPenn CDB PennSysPharmATT Univ. of Pennsylvania Department of Cancer Biology Children's Hospital UPenn Dept of Biochem & Biophys Penn Medicine E. John Wherry Berger Lab Junwei Shi Research Highlight: Delivering genome editing tools to primary cells, via Nature Reviews Drug Discovery nature.com/articles/d4157…
Congratulations to Zhendong Cao Zhendong (Tom) Cao who was a member of the Shi lab Junwei Shi


Super excited to share our work on the differential roles of distinct versions of SWI/SNF chromatin remodelling complexes in CD8 T cell exhaustion! A huge team effort from labs of E. John Wherry , Martha Jordan and Junwei Shi. doi.org/10.1016/j.immu…

It was exciting to review Liau Lab’s cutting-edge use of CRISPR tiling screens to uncover drug addiction mutations & a crucial lymphoma epigenetic window. Immensely grateful to Kohli Lab & Junwei Shi for their invaluable guidance and engaging discussions! sciencedirect.com/science/articl…

Congrats! Dr. Amy Baxter. It was such a fantastic experience to work together with you on this work.


📢Direct Enzymatic Sequencing of 5mC at Single-Base Resolution - out today in Nature Chemical Biology 👉nature.com/articles/s4158… Kohli Lab and lead author Tong Wang, MD, PhD discuss a new method, DM-seq, to specifically map ONLY 5mC through enzymatic approaches #genome #sequencing #innovations


Paper day! This one is a fantastic joint effort from Alyna Katti and Adrián Vega, PhD. I tweeted about this >1 year ago when the #preprint dropped, but there was a lot added in review, so... here is iBE - a new inducible #baseediting mouse model! 1/15 go.nature.com/3KzCo2i




In vivo perturb-seq in cancer and immune cells with John Liu, MD PhD and RaleighLab. doi.org/10.1101/2023.0… Excited for this kind of approach in adult tissues and diseases! Arc Institute UC San Francisco

Welcome to our newest Univ. of Pennsylvania Department of Cancer Biology faculty member Dana Silverbush Dana Silverbush🎗️ whose lab will harness multi-omics, machine learning, & computational tools to dissect tumor biology to improve therapeutic opportunities.

Excited to share our (with LukeGilbert) new preprint (is.gd/5kgWsI) on a new combinatorial functional genomics platform that enables 6-plex CRISPRi pooled screens. This platform works for multi-gene targeting and is especially suited for multi-enhancer targeting.



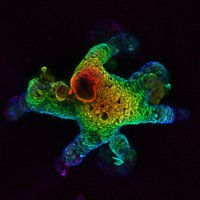
The last lead author article from my PhD is out! In Nucleic Acids Res, we report our mechanistic study on how Cas9 and AID enzymes collaborate to shape base editing outcomes. Leveraging these insights, we create hyperactive BEs with the potential for targeted gene diversification. 1/