
josh
@joshfasky
postdoc in bethesda; ☕️, read📜, ☕️, compute, 🧠, ☕️, jot📜, rinse & ♻️; @iubloomington hoosier and @usc trojan; @spursofficial; rt ≠ endorse yadda yada
ID: 285380882
https://faskowit.github.io/ 21-04-2011 01:53:42
1,1K Tweet
660 Followers
761 Following


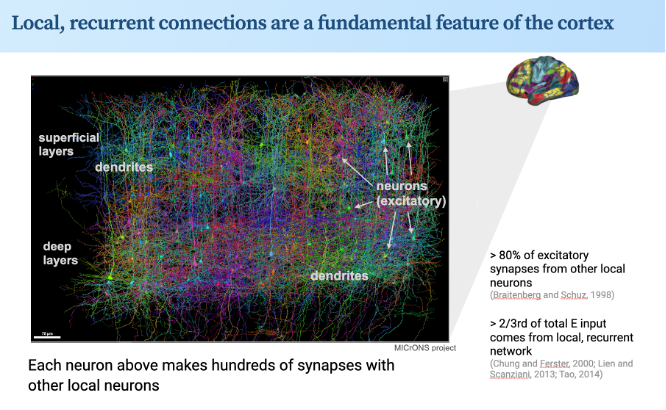
New preprint, on 'sequence filtering'. Led by Ciana Deveau, Z Zhou. We see this as a key step forw on how cortex works. All cortical areas have dense exc-exc recurrent connectivity. What do these connections do, esp in sensory ctx? Our data say: they do dynamics/time. 1/4

conn2res: a toolbox for connectome-based reservoir computing | doi.org/10.1038/s41467… Computational phenotyping of neural wiring: convert biological networks to artificial networks. by Laura Suarez ⤵️



2 years into making, the last chapter of my PhD thesis, and a set of wonderful collaborators: @m00rcheh, @ShreyDixitAI, Bratislav Misic, @caioseguin, @GZamora_Lopez, Claus C Hilgetag. 🧵 What can we learn about communication in brain networks from 7000 million virtual lesions:


I'm so excited to share the culmination of this thrilling adventure with Amy Janes and Blaise Frederick, out now in Nature Human Behaviour 📜 includes tools to detect and remove this pernicious 🎈artifact that hides in plain sight in our #fMRI datasets rdcu.be/dLgVM 🧵..


New banger from Brandon Munn and co. TL/DR: coarse-graining multi-species calcium data allows us to link across scales and make some inferences about how information processing might work in the brain. Super fun collab with Eli Muller Michael Breakspear & Ethan Scott

Arnaud, Mark and Claus C Hilgetag found that you can threshold most of the connectome, binarize the rest, and still get the same rough dynamics. It gets even more interesting because you can do the same with ANNs and get the same performance with a lot less compute time 🤌


🧠This research by Jingnan Du et al. aims to unravel the mysteries of cerebral networks through intensive #fMRI data sampling revealed a fascinating hierarchical organization within individuals. 🔓ow.ly/zwUS50SuwOB #OpenAccess Lauren DiNicola #AssociationCortex


Our cortical surface template, onavg (short for OpenNeuro Average), is published in Nature Methods. Onavg evenly samples different parts of the cerebral cortex, leading to improved performance of various analysis and reduced computational time. Guo Jiahui Ida Gobbini Haxby Lab




Our recommendations for assessing SC-based predictions of an individual's FC Individual predictions can be clouded by group-average effects Ye Ella Tian Thomas Yeo T Sarwar & Y Liu Out in Network Neuroscience: direct.mit.edu/netn/article/d…




🗣️Language is widely distributed throughout the brain🧠 In a recent correspondence in Nature Rev Neurosci, we suggest that there is no 'language network' in the brain. What appears as such is an inevitable illusion created in part by the methods we use. nature.com/articles/s4158…


First tweet on an exciting new manuscript online Nature Neuroscience - in collab with Lucina Uddin and Catie Chang. We take a fresh look at the physiological dynamics associated with the global signal 🧠... #neuroscience #neuroimaging #fMRI Read here: rdcu.be/ek01F


📰Our new research on how and when the infant human brain becomes synergistic is out now in Communications Biology! In this work, we used information theory to analyse dense EEG from preterm infants recorded during 33-45 weeks. nature.com/articles/s4200… Key findings are in the thread⬇️


