Ivan Aksamentov
@ivan_aksamentov
ID: 3131829784
03-04-2015 22:38:44
72 Tweet
314 Followers
1,1K Following



A *huge* thank you to Ivan Aksamentov for implementing this feature, and to Tony Qu for suggesting this & for translating everything into Chinese! 🙏🏻🇨🇳 We hope this makes CoVariants.org even more accessible to people around the world!🌏 2/5




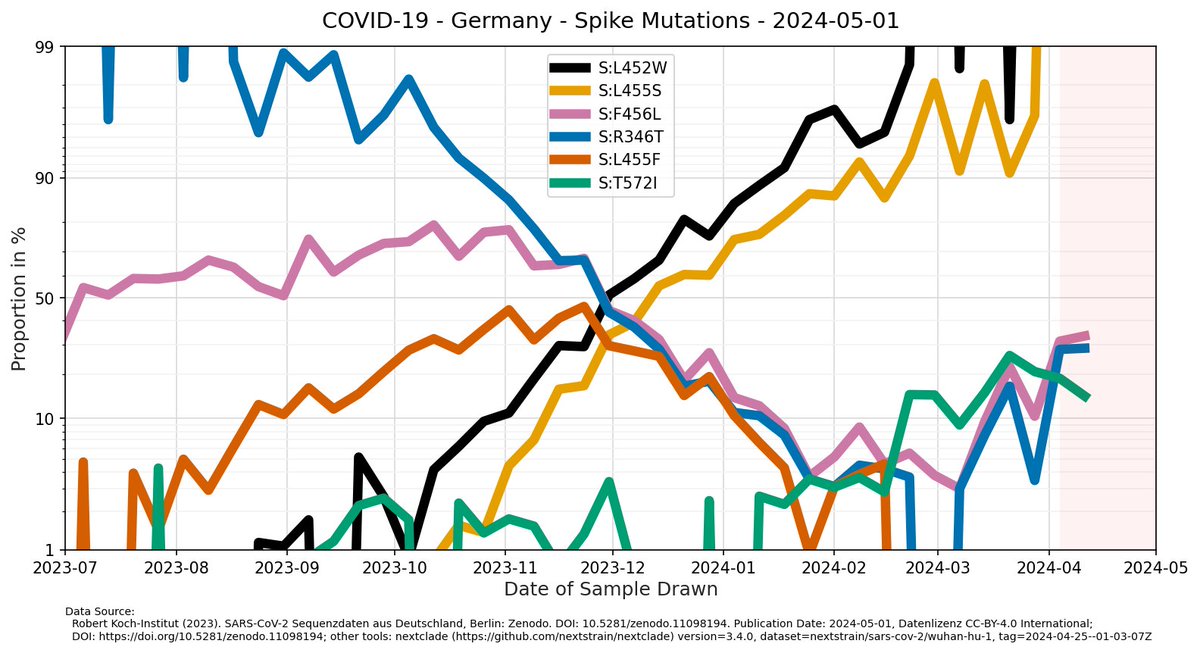
Mike Honey --Neon-Newton-- As sample size reduced to 20.. 25 samples per week, it's a bit...noisy 🤪🙈 My analysis with nextclade of RKI sequence data in the graphs below (x-axis: date of sample drawing/swab). So pretty likely we hit 50% around last week to now here in Germany.








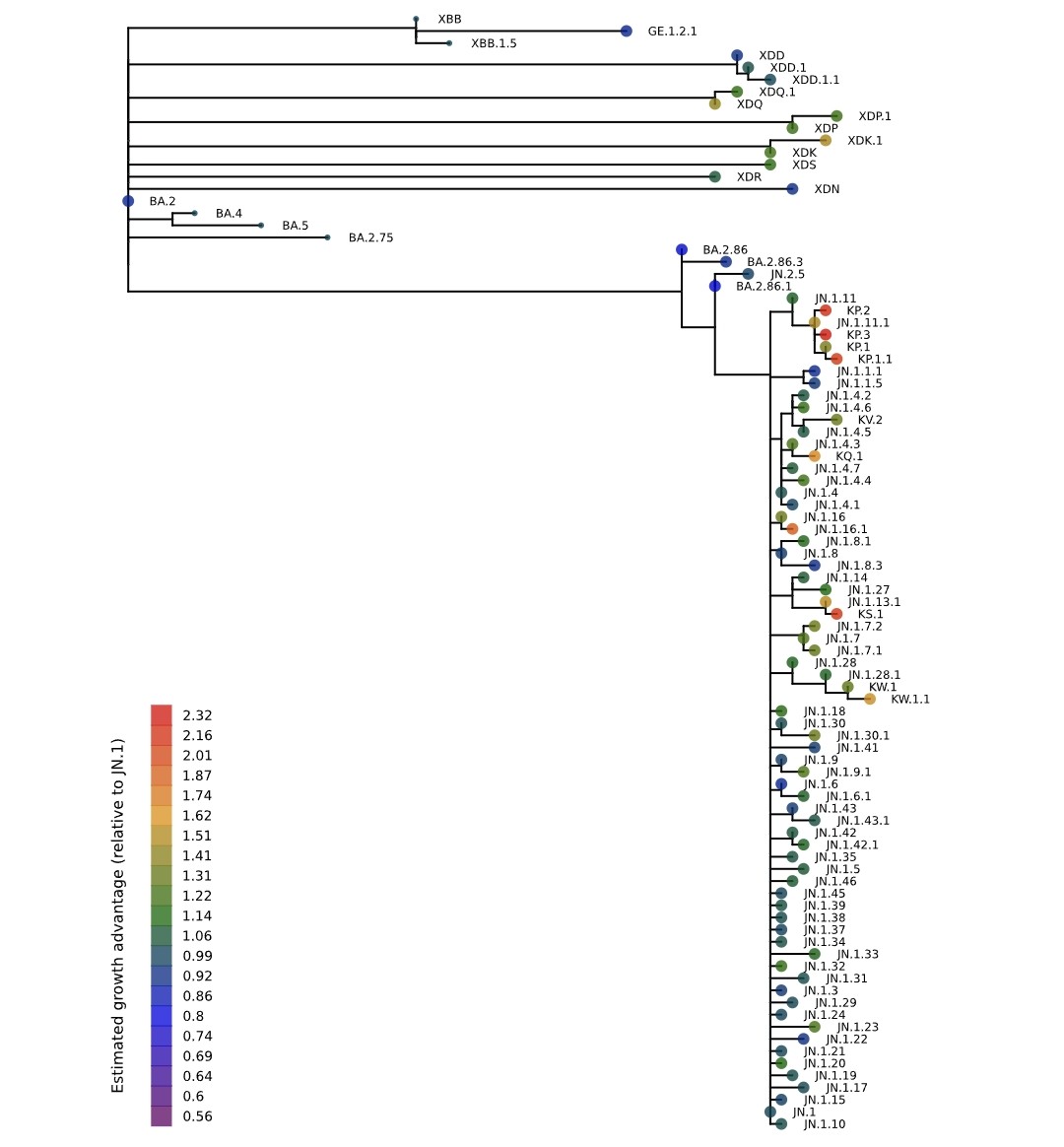
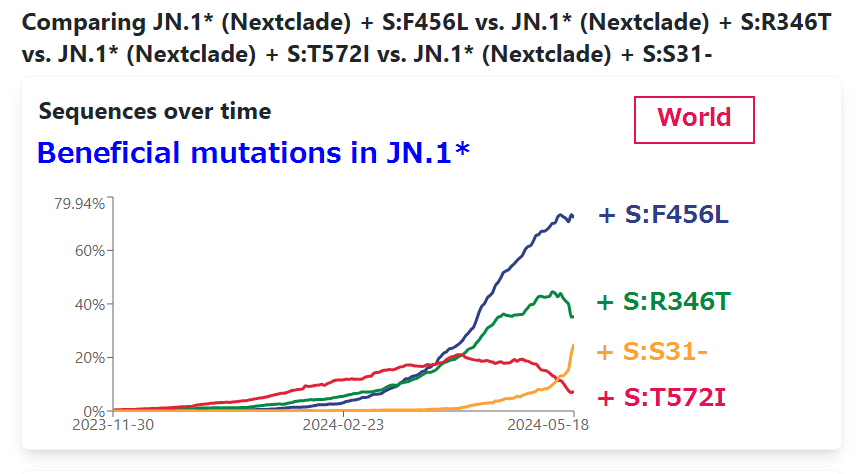
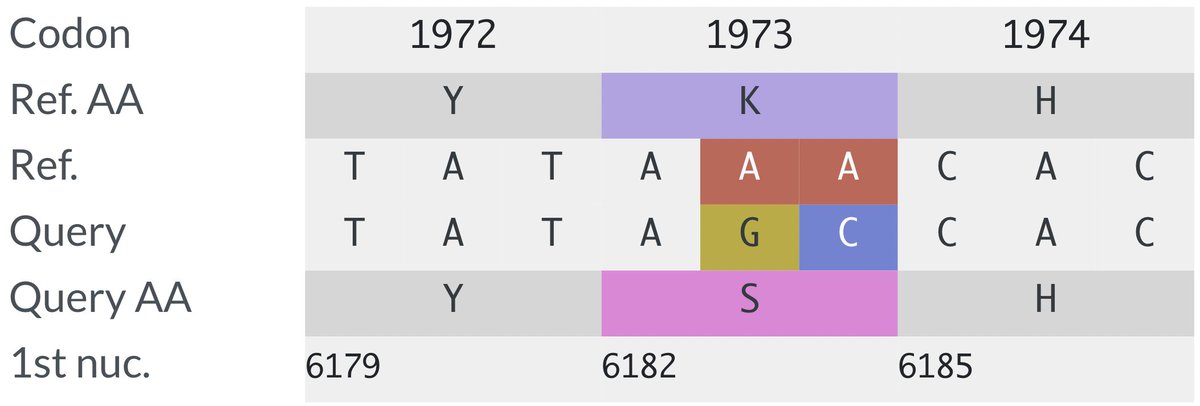
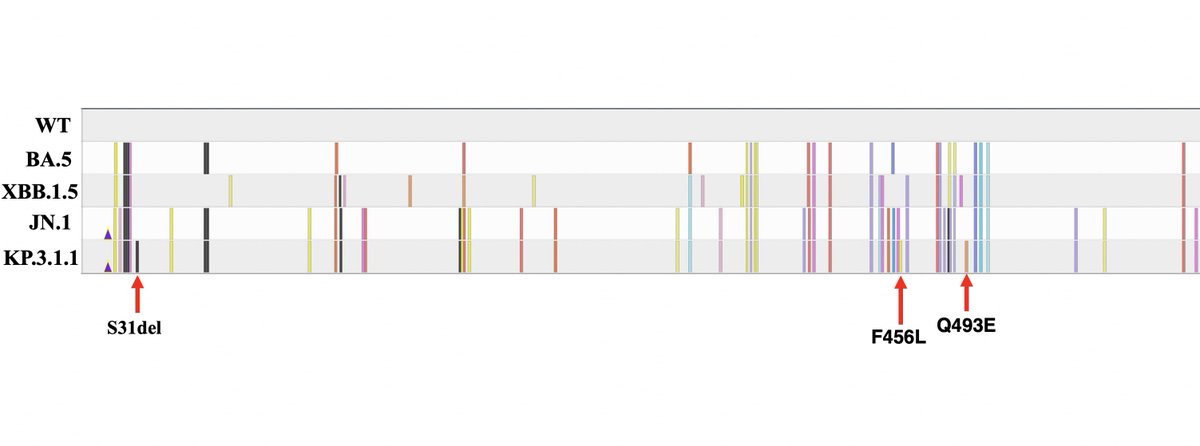
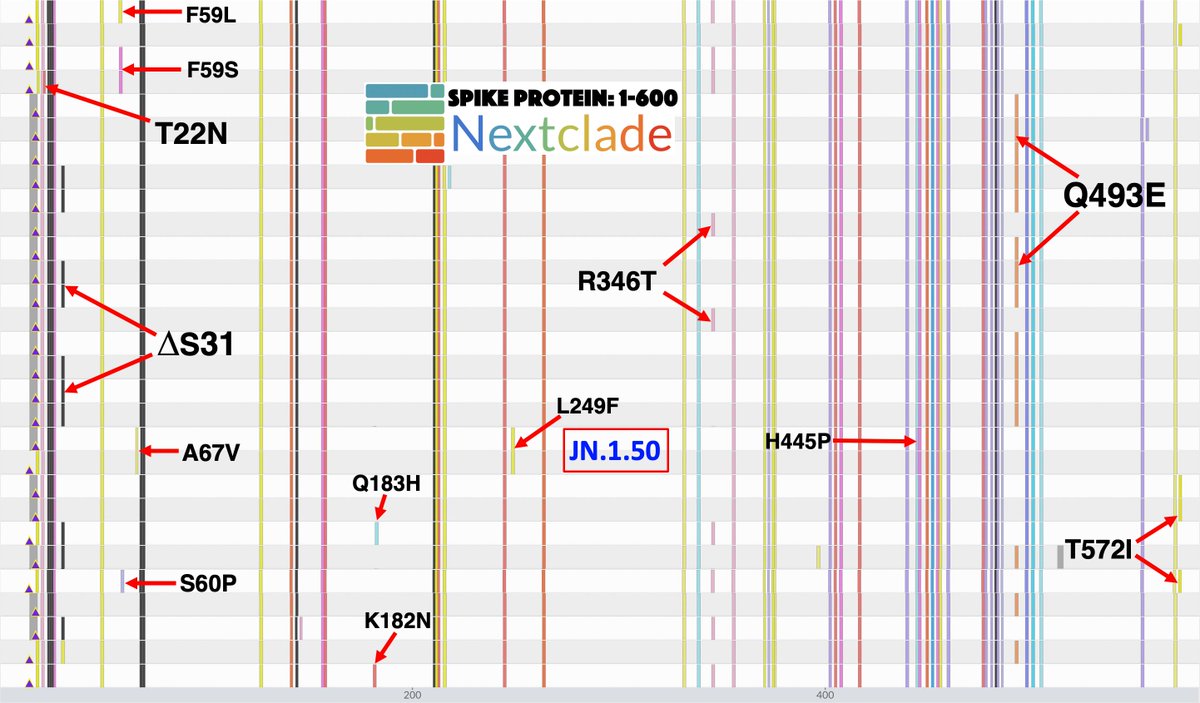
. Ben Murrell is doing the best variant growth modeling in the world, & his latest results confirm most of what we've thought: KP.3 is the fastest large variant, & its sublineage KP.3.1.1—w/the highly advantageous, glycan-creating S:∆S31—is easily the fastest in the world. 1/15