
Marcus Hartmann
@hartmannlab
Structural Biochemistry @MPI_Bio & @UNI_TUE
Understanding and Manipulating Molecular Machines
ID: 1272297370663956482
http://www.eb.mpg.de/protein-evolution/marcus-hartmann 14-06-2020 22:38:35
139 Tweet
176 Followers
95 Following

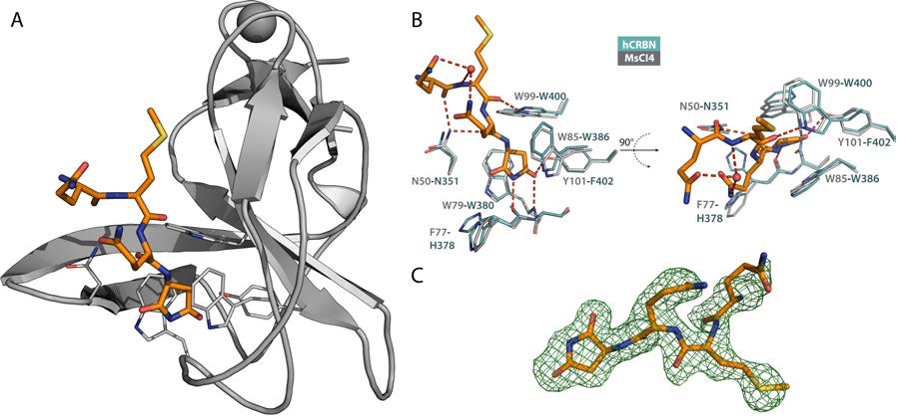
We couldn't agree more - this is a fascinating topic and we are more than happy to provide the structural basis for the recognition of the C-terminal cyclic imide #degron in our new communication Biochemical & Biophysical Research Communications: sciencedirect.com/science/articl…

🥳🥳 Congrats, great job! 🥳🥳 Fantastic PhD defense by Joe this week, showcasing great results from your collaborative salmonella anti-virulence project with @samuwag Antti Poso Brönstrup Lab! #Salmonella, beware of Joe! TIPP/IMPRS PhD Reps @MPI_Bio, FML & @uni_tue



🚨PhD alert just before Xmas 🎅 My lab offers a PhD position within the @GRK2873 funded by DFG public | @[email protected] The project is devoted to #PROTACs and E3 ligand #chemistry #lifesciences For more information visit our website 👇👇 and apply! Please help & retweet!

🧪Happy to share our work on three-branched CRBN ligands for MG and #PROTACs design. A multicomponent reaction as synthetic entry🤯Nice work by my PhD student Robert and Krönke Lab Marcus Hartmann at the interface between Chemistry & Biology. #OpenAccess rsc.li/3wfEtJg

Happy to share the missing link between the action of #IMiDs & #CELMoDs and the natural #CRBN degron Biochemical & Biophysical Research Communications: Neo-substrates mimic every single H-bond of cyclic imide #degron recognition, inevitably inheriting sequence-independence of natural #degrons 👇 sciencedirect.com/science/articl…

After long time in oven, happy to share w/ Marcus Hartmann PNASNews our work #Cyanobacteria #Carbon #Sensor SbtB possesses atypical #Redox regulated #Apyrase (hydrolysing ATP/ADP) to control HCO3 transport Generous fund by Tübingen University Excellence Projects Controlling Microbes to Fight Infections @MPI_Bio pnas.org/doi/10.1073/pn…

An excellent perspective by authors from AstraZeneca about fragment-based approaches to identify ligands for E3 ligases. Of especially high relevance for the field of targeted protein degradation (#TPD). ACS Publications Bio & Med Chem Content pubs.acs.org/doi/10.1021/ac…

Check out our work on regulating #Salmonella virulence bioRxiv! Great job by Joe, Wieland, Thales, BangeBalcony, Antti Poso & @samuwag: HilE represses the activity of HilD via a mechanism distinct from that of intestinal long-chain fatty acids. biorxiv.org/content/10.110…

Congrats, truly seminal work! Since our initial discovery of bacterial histones, we too worked on the very same histone protein & now present its crystal structure & DNA-binding properties. Great work by Yimin, Birte & collaborators Marcus Hartmann @MPI_Bio doi.org/10.1101/2023.0…



A press release about our recent paper in PNASNews pnas.org/doi/10.1073/pn… Cyanobacterial protein integrates the circadian clock information growkudos.com/publications/1… via @growkudos Tübingen University Excellence Projects Controlling Microbes to Fight Infections @MPI_Bio

Great to see this out! Enormous MedChem efforts led to new SAR aspects of VHL ligands 🧪💊 #TPD bigs thanks to Alessio Ciulli +team & Izidor Sosič +team and colleagues from Rheinische Friedrich-Wilhelms-Universität Bonn


It’s #FluorescenceFriday so some #RealTimeChem from my lab: purification (until battery crashed) of a fluorescently labelled non-chiral, non-glutarimide/uracile CRBN ligand with outstanding affinity. #TPD tweeps, it will blow your mind 🤯 next step: in vitro assays Marcus Hartmann

Sleeves up for infection research: Our postdoc Joe Joiner researches anti-infectives to prevent #Salmonella infection. He’s not only appreciated as a talented researcher, but also as a great teacher by his students! Postdoc Appreciation Week Germany #PostDocAppreciationWeek @MPI_Bio






