Matthieu Gagnon
@gagnon_lab
Associate Professor, Microbiology & Immunology/Biochemistry & Molecular Biology, Structural studies of translation machinery, Ribosome, Antibiotics, Cryo-EM
ID: 1687144263535009810
03-08-2023 16:52:09
113 Tweet
118 Followers
111 Following

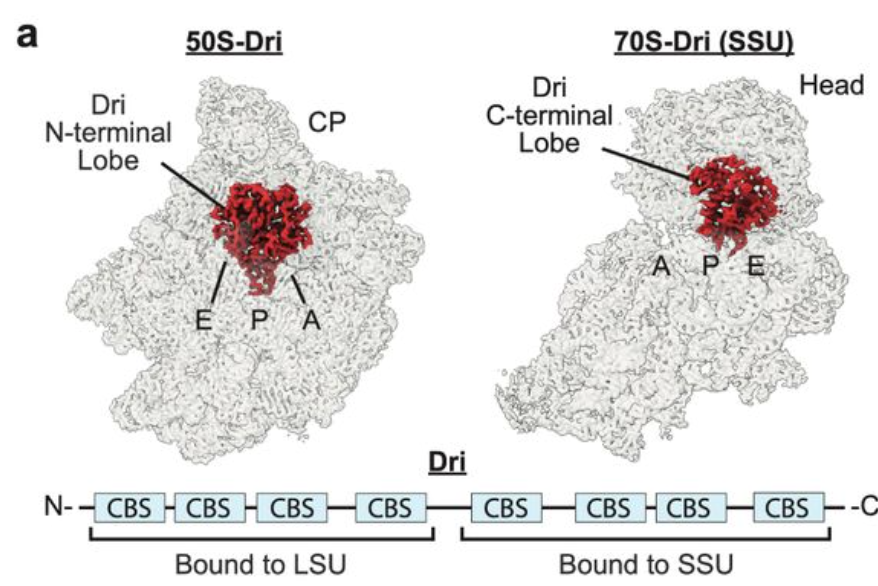
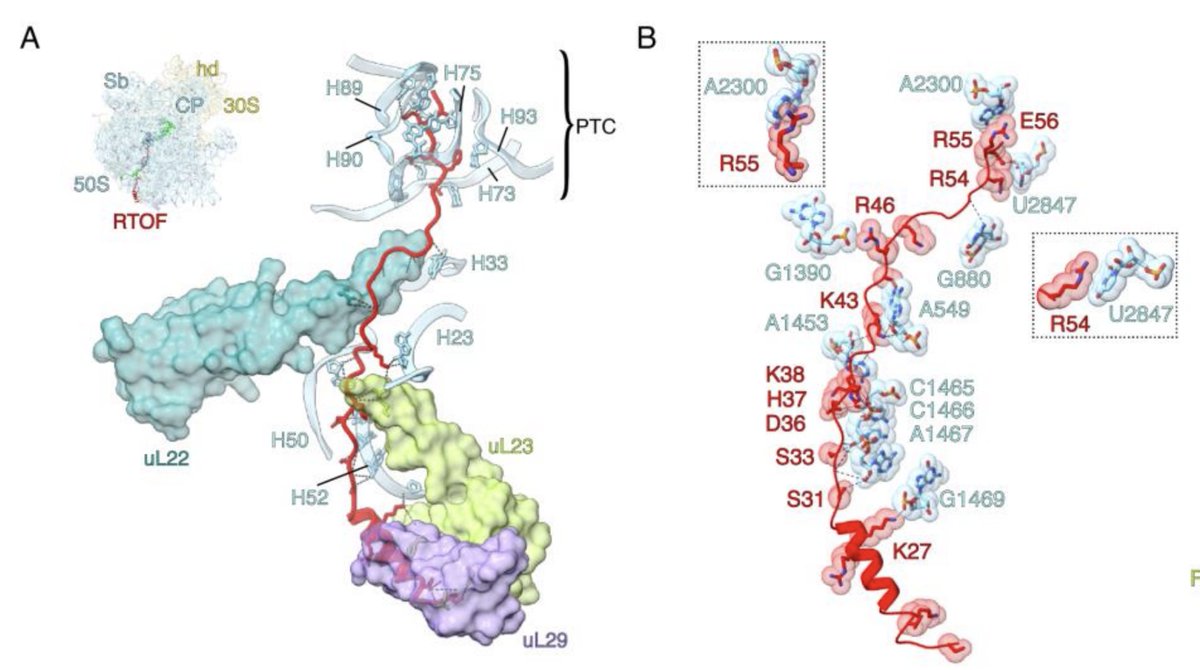
In our new study we reveal how NAC coordinates and regulates successive co-translational nascent chain ⛓️ processing on the mammalian ribosome out today in nature rdcu.be/dRAQY collab with Deuerling Lab Shu-ou Shan Caltech Swiss National Science Foundation European Research Council (ERC) ETH Zurich

Principal Investigator Matthieu Gagnon and researchers at UTMB Health use cryogenic-electron microscopy to elucidate the mechanism of GDP dissociation from RF3 catalyzed by the Escherichia coli 70S-TC. buff.ly/4dxEqwm

The beauty of protein synthesis and its main players - ribosomes, mRNA and protein, seen in artistic view at CSHL. Once again at CSHL Meetings Translational control and protein synthesis.



Now out in peer-reviewed format at Nature Chemical Biology …rdcu.be/dXhoi




We are excited to share our latest work on the cooperation between RNA polymerase and the ribosome in bacteria: science.org/doi/10.1126/sc… The work was supported by IGBMC, Inserm, CNRS 🌍, -, @ERC_research, IMCBio Graduate School, and Instruct-ERIC. 1/7


Out just now at Nature Communications: using cryo-EM and cryo-electron tomography, we provide evidence that structurally heterogeneous ribosomes can cooperate in protein synthesis in bacterial cells: nature.com/articles/s4146…



1 of 2: Ever wondered how newly synthesized proteins are released from the ribosome? An exciting collaboration between the Polikanov and Matthieu Gagnon groups elucidates this conundrum.


Preprint from our collaboration with the labs of Gerry Wright and Shura Mankin Mankin_Vazquez_Lab identifying the first translation inhibitor binding to the E-site of the bacterial ribosome… researchsquare.com/article/rs-692…