
Yilei Fu
@fuyilei96
Postdoc @sedlazeck lab at HGSC, BCM. Ph.D @RiceCompSci, Treangen Lab
ID: 822547192464424960
https://fu-yilei.github.io/ 20-01-2017 20:51:47
293 Tweet
136 Followers
389 Following

I'm sharing a public version of my talk slides CGSI-The Computational Genomics Summer Institute two weeks ago - an update from my #LLMs #compbio Primer talk last year - w/ a focus on genomics FMs (DNA, scRNA). As we're exploring related topics, we remain very interested but also skeptical. 1/3 cs.cmu.edu/~jianma/talks/…

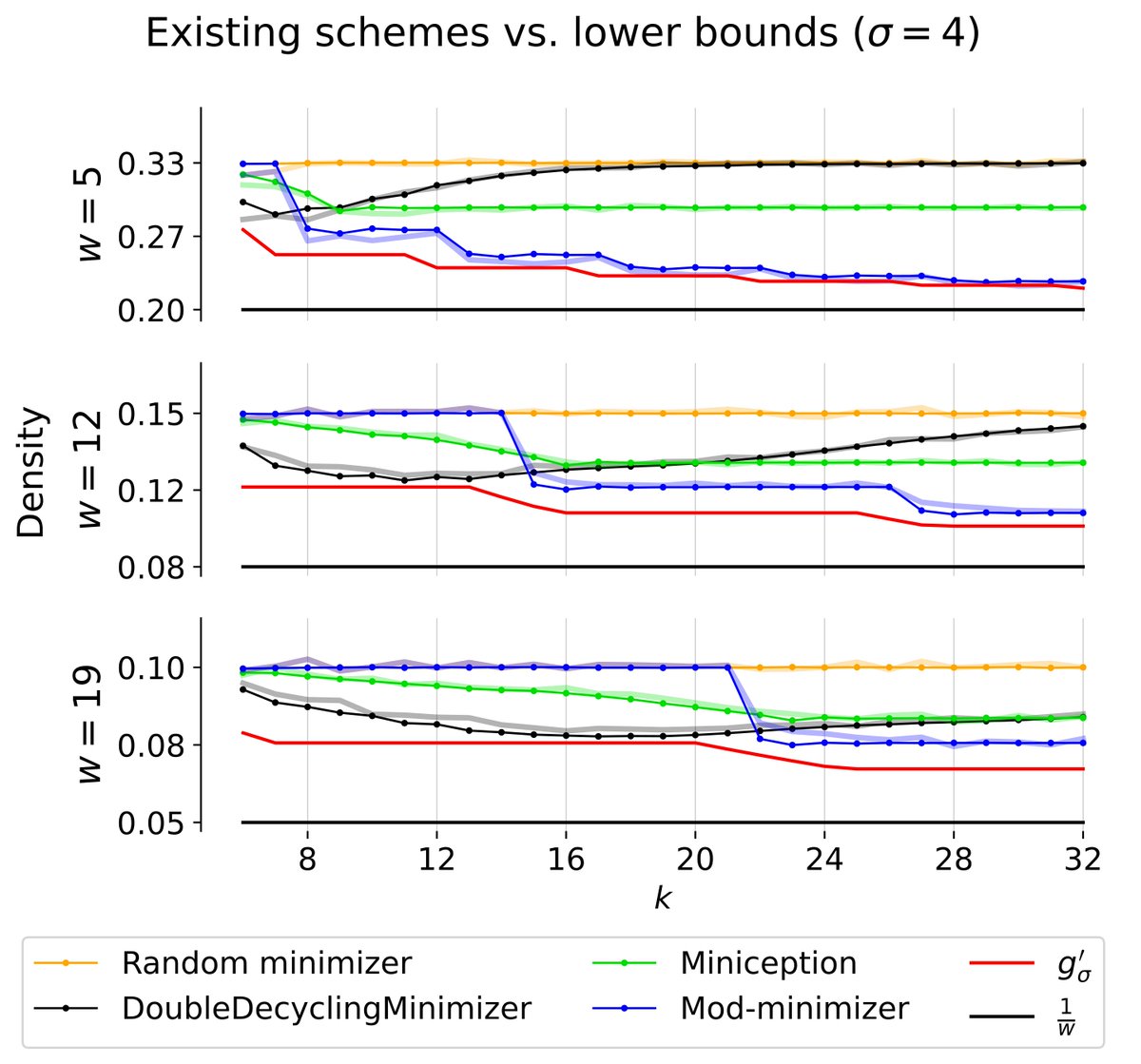
In a new preprint, Ragnar {Groot Koerkamp} 🦋 and I led the derivation of a new lower bound, g’ (red), on k-mer sampling scheme density: - our bound appears to be tight for k ≡ 1 (mod w). - existing schemes are nearly optimal! - mod-minimizers are optimal for large sigma for k ≡ 1 (mod w)

How to annotated SV with pop. frequency? Using PacBio & Oxford Nanopore! Gnomad 32% & STIX: 95.9% of SV from Genome in a Bottle annotatable! Huge implications in SV filtering for eg: GREGoR Consortium & #cancer & #disease studies! Lead by xinchang zheng & ryanlayer biorxiv.org/content/10.110…




We summarized state-of-the-art computational methods for DNA methylation analysis using long-read sequencing, covering everything from base calling to sample-level, cell-type-level, and even population-scale analysis. Huge Thanks to Fritz Sedlazeck and Winston Timp for this great work!

Long reads Oxford Nanopore PacBio can provide insights into epigenetics. We (Yilei Fu Winston Timp) summarize the latest #Bioinformatics approaches to analyze methylation & why this matters! Out now Nature Reviews Genetics bit.ly/4cjxUtl BCM HGSC BCM Department of Molecular and Human Genetics Rice Computer Science Johns Hopkins University


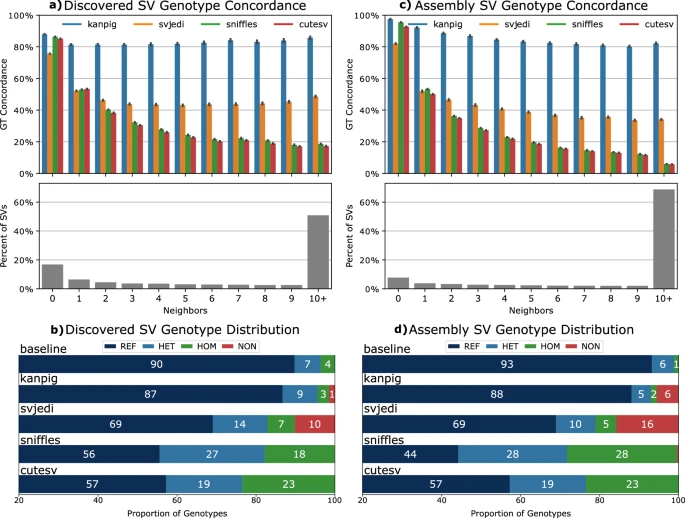
How to accurately genotype SV is important & we discovered a huge bias in other methods! Many improvements across population wide SV calling! Out now Kanpig: nature.com/articles/s4146… Nature Communications Great work from BCM HGSC Broad Institute #Bioinformatics

Come join us at our 7th #bioinformatics #hackathon BCM HGSC From the Labs at Baylor College of Medicine around Structural Variants, Graph genomes & many related topics: fritzsedlazeck.github.io/blog/2025/hack… Groups will work on interesting topics that will be published in F1000. Registration closes 10th of Aug!

1/ 🧬 A Hitchhiker’s Guide to Long-Read Genomic Analysis is out now Genome Research! This mini-review walks through the latest advances in long-read DNA sequencing — from assemblies to variant calling to epigenetics. Link 🔗 genome.cshlp.org/content/35/4/5… 🧵👇


Initial release of Pomfret for methylation assisted phase block joining for more contiguous phased assemblies without any additional library prep from Oxford Nanopore data. Such amazing work from Xiaowen Feng ! #NanoporeConf github.com/nanoporetech/p…



🎉 Congratulations to our Early Career Researcher Pilot and Feasibility Awardees! 👏 Yilei Fu – Fritz Sedlazeck 👏 Cheuk-Ting Law – Kathleen Helen Burns 👏 Yang "Young" Zhang – Zong Lab 👏 Elliot Swanson – Stergachis Lab 👏 Andy Russell – Fei Chen Lab 👏 Hang Su – Broad Institute



Come join us on our SV, graph genome hackathon August 27–29, 2025: fritzsedlazeck.github.io/blog/2025/hack… Seats are filling up! It will be great opportunity to work together on some prototypes. We will publish them together in F1000. Rice University BCMHouston #bioinformatics #openscience







