Fabien Plisson
@fabienplisson
Drug Discovery, ML+AI & Peptide Design | Rosenkranz Award 2021 | 🇫🇷 🇦🇺 🇲🇽 | Dad | Founding ingeniebio.com ORCID 0000-0003-224
ID: 2904831962
http://plissonf.github.io 20-11-2014 05:27:07
2,2K Tweet
1,1K Followers
1,1K Following

Remember Golden Gate Claude? Etowah Adams and I have been working on applying the same mechanistic interpretability techniques to protein language models. We found lots of features and they’re... pretty weird? 🧵


Brilliant video by Veritasium on the development of protein structure prediction featuring #AF2 and #Rosetta series with David Baker youtu.be/P_fHJIYENdI?si…



Half of an AI scientist is rejecting or accepting hypotheses. FutureHouse and ScienceMachine just put out ~300 novel hypotheses from ~50 published papers along with ground-truth data. Humans take 4.2 hours to solve these and frontier models get 10-20% correct. SWE-bench for bio



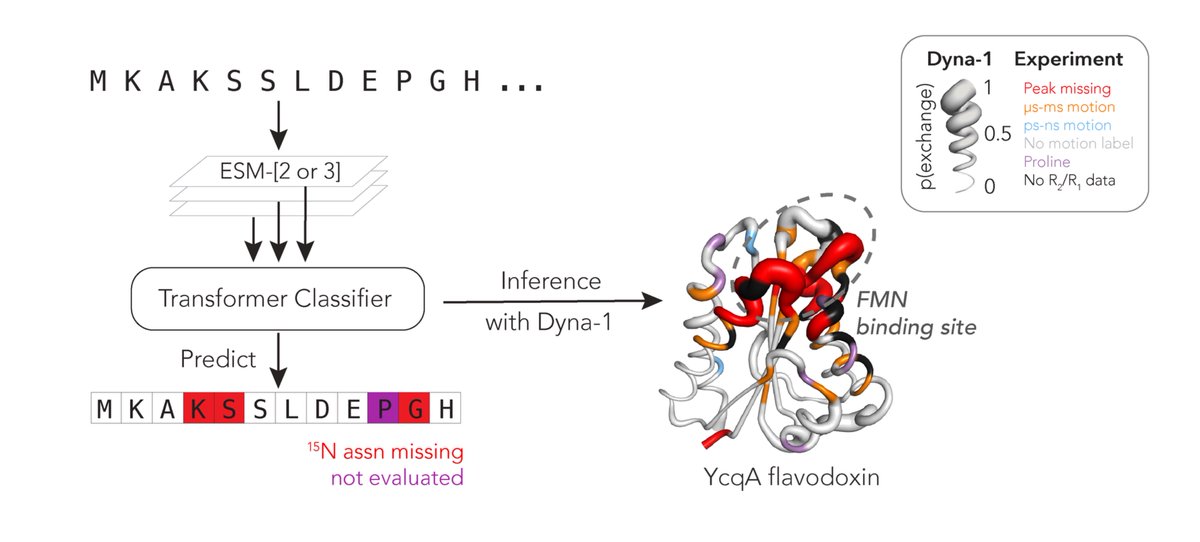
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵






Ever wondered how to spot new antibiotics hiding in biological data? Our latest paper in Nature Protocols Nature Portfolio offers a practical, end-to-end ML playbook for mining genomes & proteomes—AI vs AMR, one dataset at a time. Link: rdcu.be/el2Go







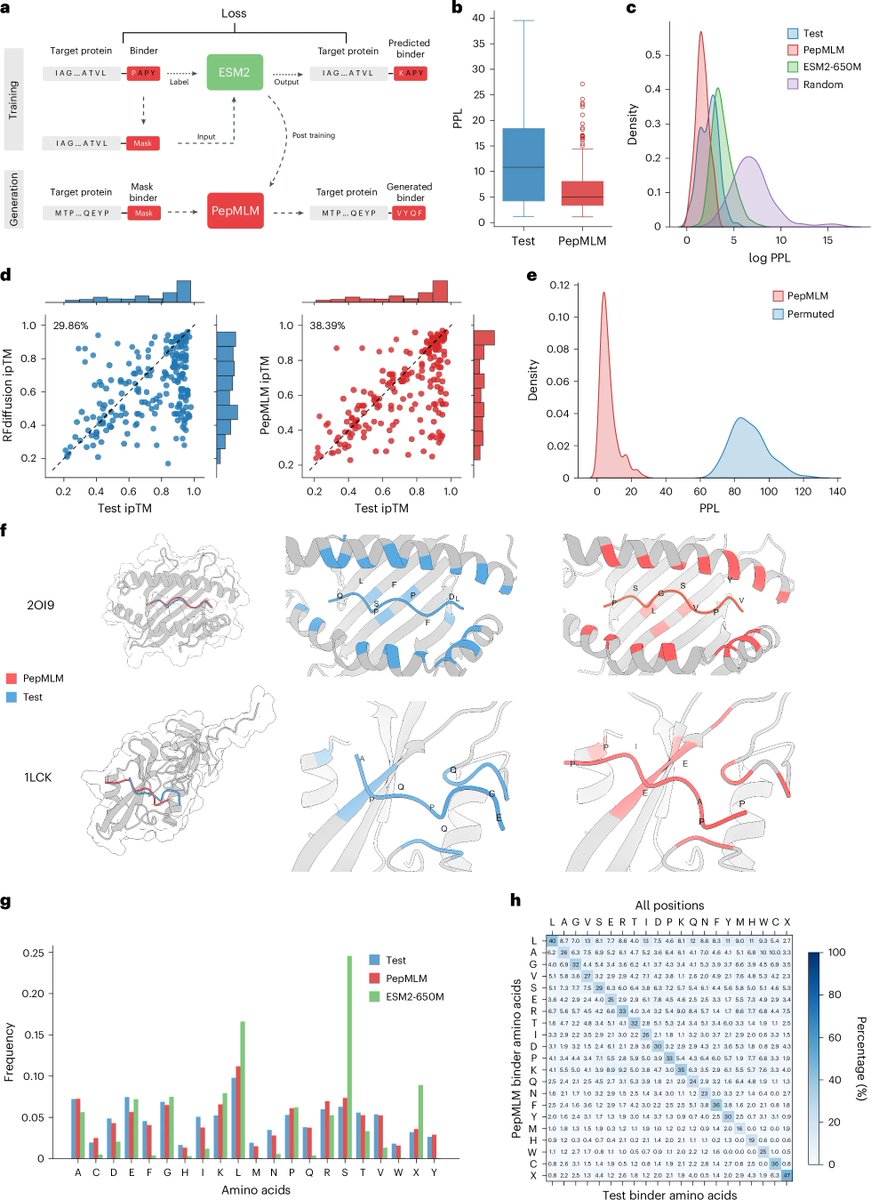
Happy to announce that PepMLM is now published in Nature Biotechnology Nature Biotechnology after 2.5 years (in the pre-RFDiffusion era 🥲) A brief recap: We simply concatenate the target protein sequence with the peptide binder sequence, mask the whole binder region, and fine-tune