
Neo Shi Yong
@drneosy
Tumor Immunologist |either in Lab or Kitchen👨🏻🍳🍜 @astar_sign @karolinskainst
ID: 202288842
13-10-2010 18:25:25
243 Tweet
112 Followers
160 Following


Excited to share our paper in Nature Biotechnology, work led by Shaobo Yang and Michal Sheffer in my lab in collaboration with Jiahe Li University of Michigan nature.com/articles/s4158… #NK #immunotherapy #bacteria Dana-Farber News Dana-Farber #RomeeLab


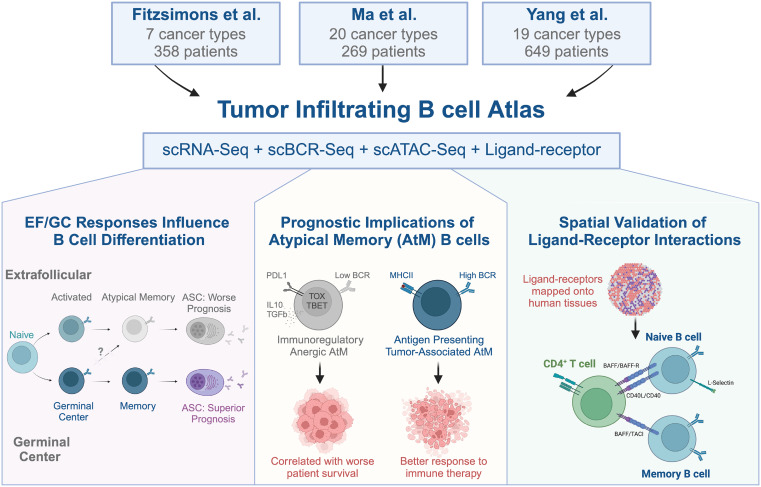
ScRNA-seq defines pan-cancer atlas of tumor-infiltrating B cell subsets Preview Cancer Cell cell.com/cancer-cell/fu… cell.com/cancer-cell/fu… doi.org/10.1016/j.cell… science.org/doi/10.1126/sc…






Comprehensive snapshots of NK cells functions, signaling, molecular mechanisms and clinical utilization Youssef Jounaidi Signal Transduction and Targeted Therapy (STTT) nature.com/articles/s4139…


Disseminated tumor seeding cells utilize the PGE2-EP2/EP4 axis to induce NK cell dysfunction and immune escape following organ colonization Cell Reports Jan Böttcher @AMPedde cell.com/cell-reports/f…


Excited to share our new paper in Journal of Experimental Medicine 🎉 We show that NK cells use autocrine TGF-β1 signalling to establish tissue residency, form a highly cytotoxic subset, and protect the tissue against viral persistence. rupress.org/jem/article/22…


New findings cast light on the role of understudied DN3 #MemoryBcells in cancer, and shows these cells correlate with progression and poor clinical outcomes in #HNSCC and melanoma. Tullia Bruno, PhD Dr.YayaTheScientist Pitt Micro/Immuno scim.ag/4if1Mc5















