Diogo Borges
@diogobor
Bioinformatics | Proteomics | Artificial Intelligence | Researcher
ID: 79576443
https://bit.ly/3rKnJ9a 03-10-2009 22:07:09
141 Tweet
216 Followers
318 Following


Very good talk by Fan Liu Fan Liu lab in #ASMS2023. If you are interested in knowing more about Scout, please visit my poster (booth MP 136) tomorrow morning.




Yay! This is what it's all about. Until next year #TeamBrazil. Hope you can all join us at #ASMS2024. Am Soc for Mass Spec #ASMS2023

Open applications for 12o. edition of Proteomics Workshop! Join us!jennifer van eyk;Andreas Mund;@ProteomicsNews;kusterlab (Inactive)@Gundrylab;@GarciaLabMS;Susan Weintraub;Diogo Borges;Brian Searle; Alexey Nesvizhskii linkedin.com/feed/update/ur…



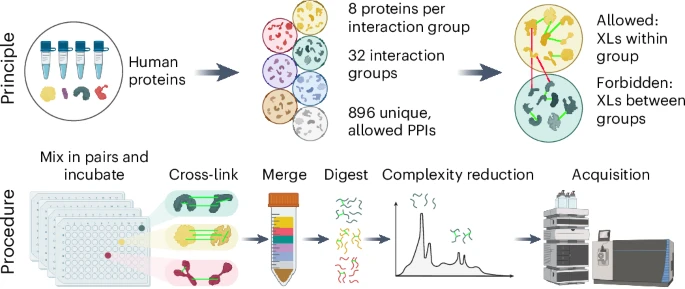
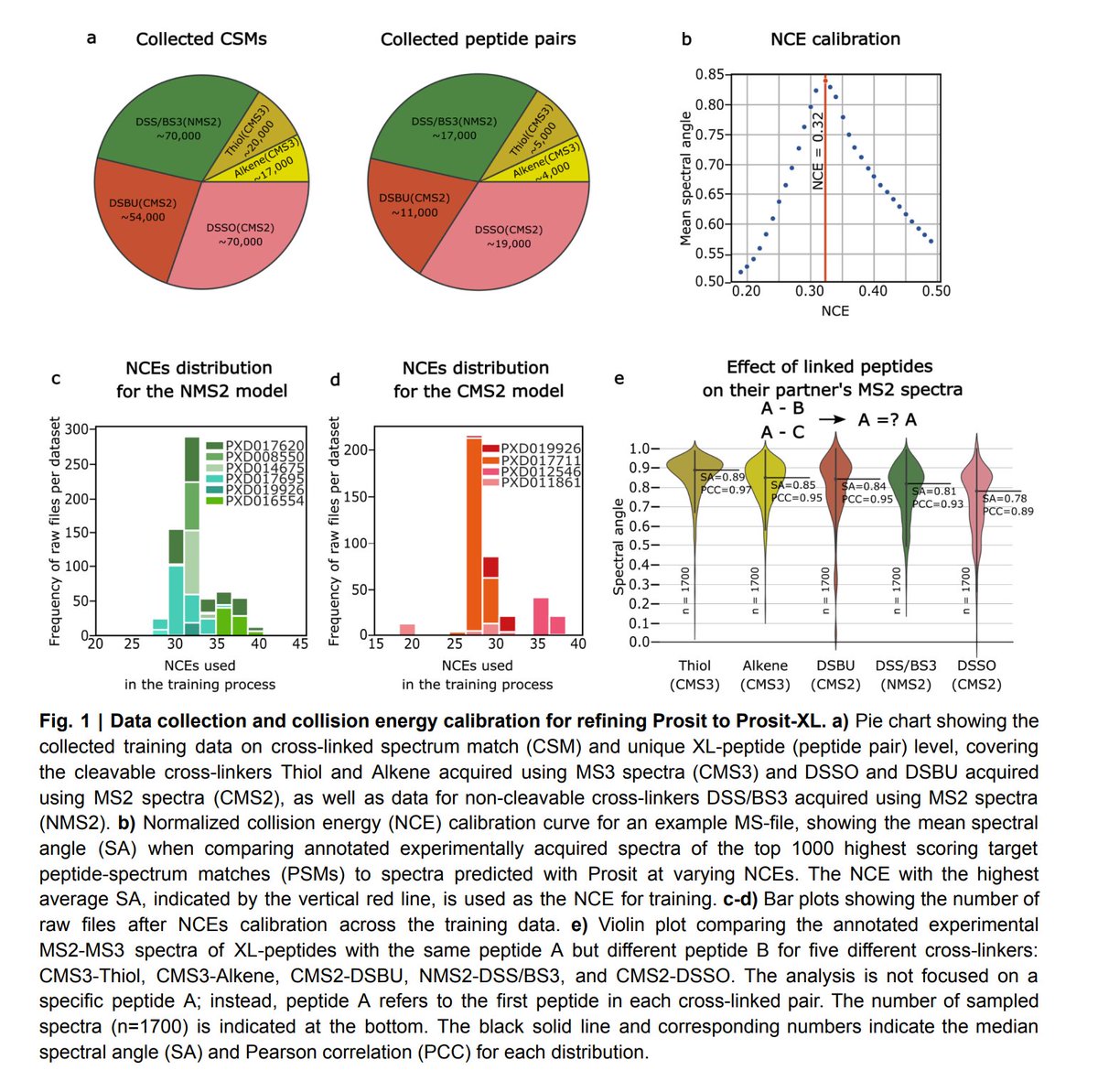
Two steps to more robust XL-MS-based interactomics: Using 100s of recombinant proteins, Milan Clasen, Max Ruwolt et al. developed proteome-scale XL-MS ground-truth standards + SCOUT, a new search engine uniting speed, sensitivity, and stringent FDR control. rb.gy/cvosfj

Our last work has been published in Journal of Proteome Research. Check it out RawVegetable 2.0, the new software for refining XL-MS data acquisition. Megamilan paulo costa carvalho pubs.acs.org/doi/10.1021/ac…





.Fan Liu lab Max Ruwolt Diogo Borges @milanzord @absea_bio @Julia_RtaAn introduce a XL-MS standard based on human proteins or protein fragments produced in E. coli. The paper also introduces Scout, a search engine for XL-MS studies with cleavable cross-linkers. nature.com/articles/s4159…