ZhangLab_SLU
@dapengzhang5
bioinformatics; comparative genomics; biological conflicts; toxin/effector systems; protein evolution; diversification mechanisms; antibiotics and resistance;
ID: 1025931866392682500
https://zhangbioinfo.weebly.com/ 05-08-2018 02:29:52
77 Tweet
93 Followers
121 Following

DeepMind open-sources AlphaFold 2 for protein structure predictions wp.me/p8wLEc-blZO by Kyle Wiggers


Joe Bondy-Denomy Same here. Mougous lab and alum used to refer to their 2012 Biology Direct (Zhang et al) paper as the bible



Another recent work where we identified the potential toxins behinds Huanglongbing and Zebra chip: Comparative Phylogenomic Analysis Reveals Evolutionary Genomic Changes and Novel Toxin Families in Endophytic Liberibacter Pathogens | Microbiology Spectrum journals.asm.org/doi/10.1128/Sp…


Look who's on the cover of Developmental Cell:the humble soil bacterium #Bacillus subtilis,in the act of sporulating,depicted using marbles and wooden blocks! Our paper tackles a ~50 yr old mystery of how the cell coordinates assembly of 2 supramolecular structures:sciencedirect.com/science/articl…


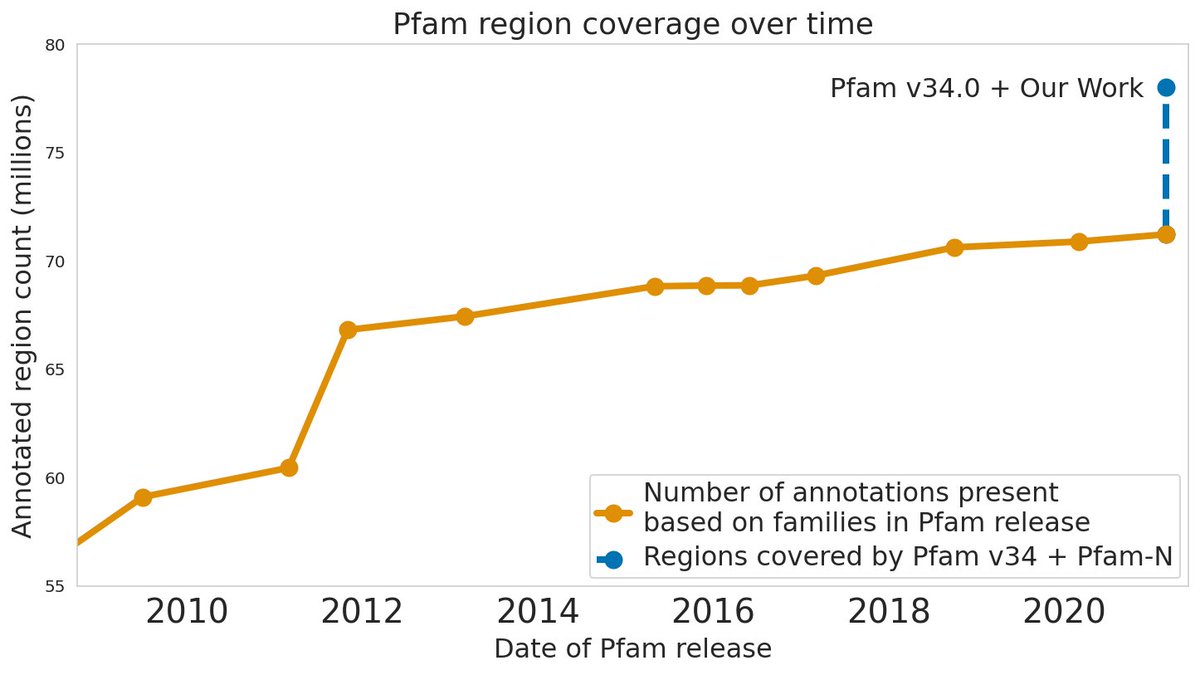
Machine learning can help read the language of life: blog.google/technology/ai/… Discussing our work in Nature Biotechnology, our interactive preprint, and why Google is involved.







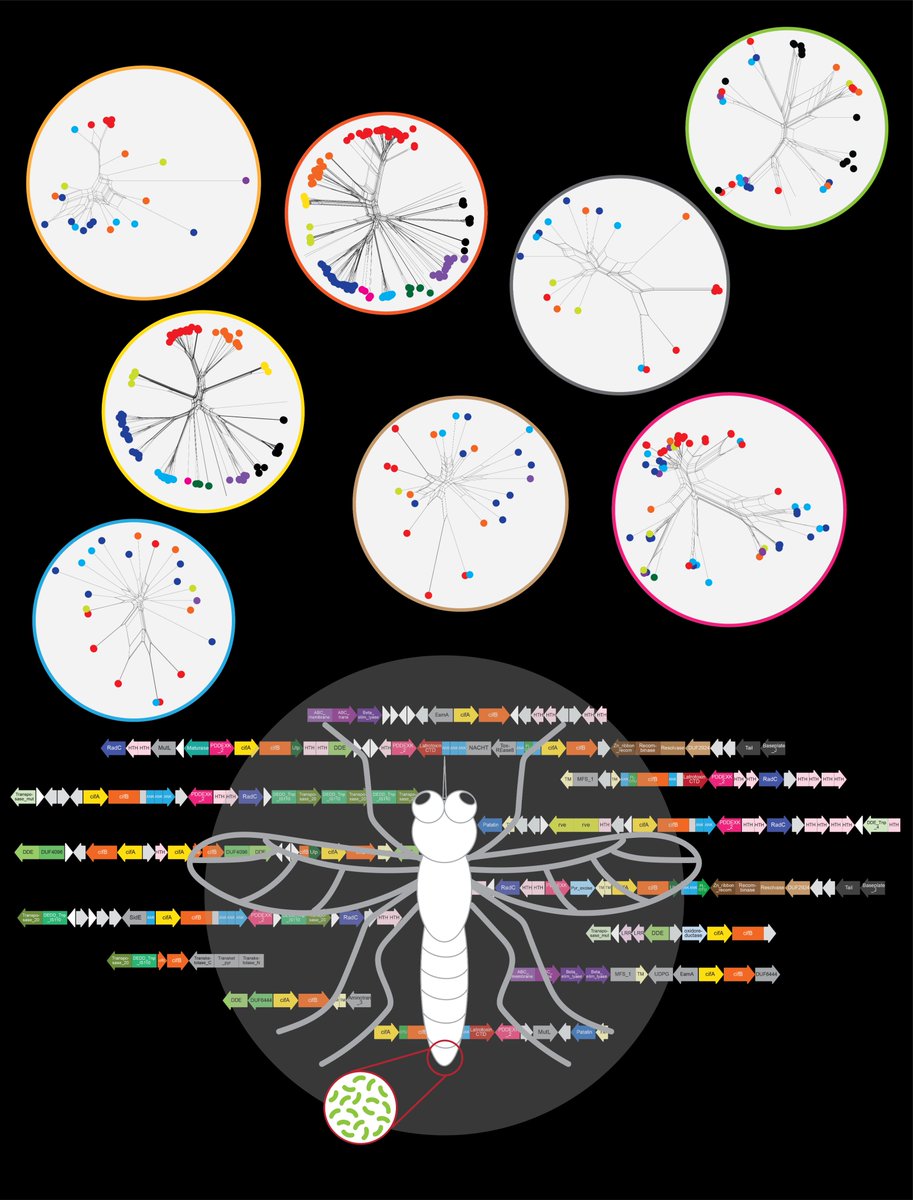
Wolbachia-mediated Cytoplasmic Incompatibility, transposons and other entanglements Our new collaborative work with ZhangLab_SLU lab is out. It investigates the fascinating phenomenon of the genomic basis of Cytoplasmic Incompatibility (CI) and its ultimate origins. This