Muhammad Munim
@mbmunim_
PhD candidate - @mvh_lab @MIT - fascinated by all things metabolism | @UofT '20 | 🇵🇰🇨🇦 he/him
ID: 1044274147134787584
24-09-2018 17:15:33
115 Tweet
172 Followers
457 Following










Recent Beamish award winner Tao Huan Nature Metabolism University of British Columbia describes how to avoid “nonsensical” results in metabolomics doi.org/10.1038/s42255…






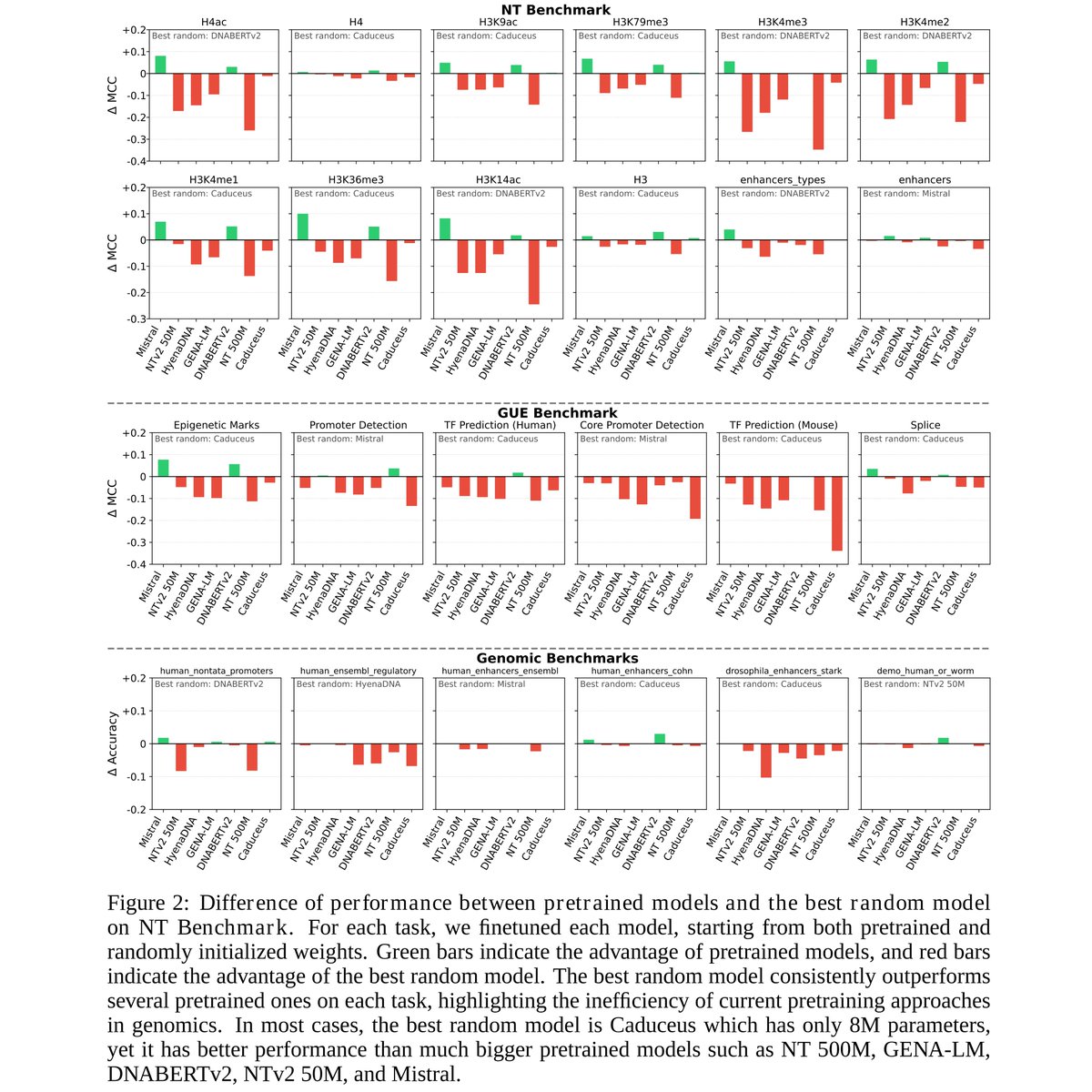
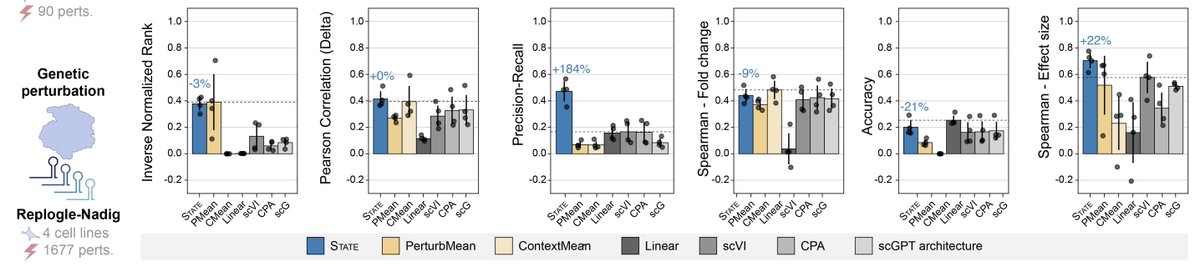
Uh oh: deep learning models in genomics lose badly to very simple linear models. Could they have been over-hyped? (new paper by Simon Anders Constantin Ahlmann-Eltze (@const-ae.bsky.social) and Wolfgang Huber) nature.com/articles/s4159…

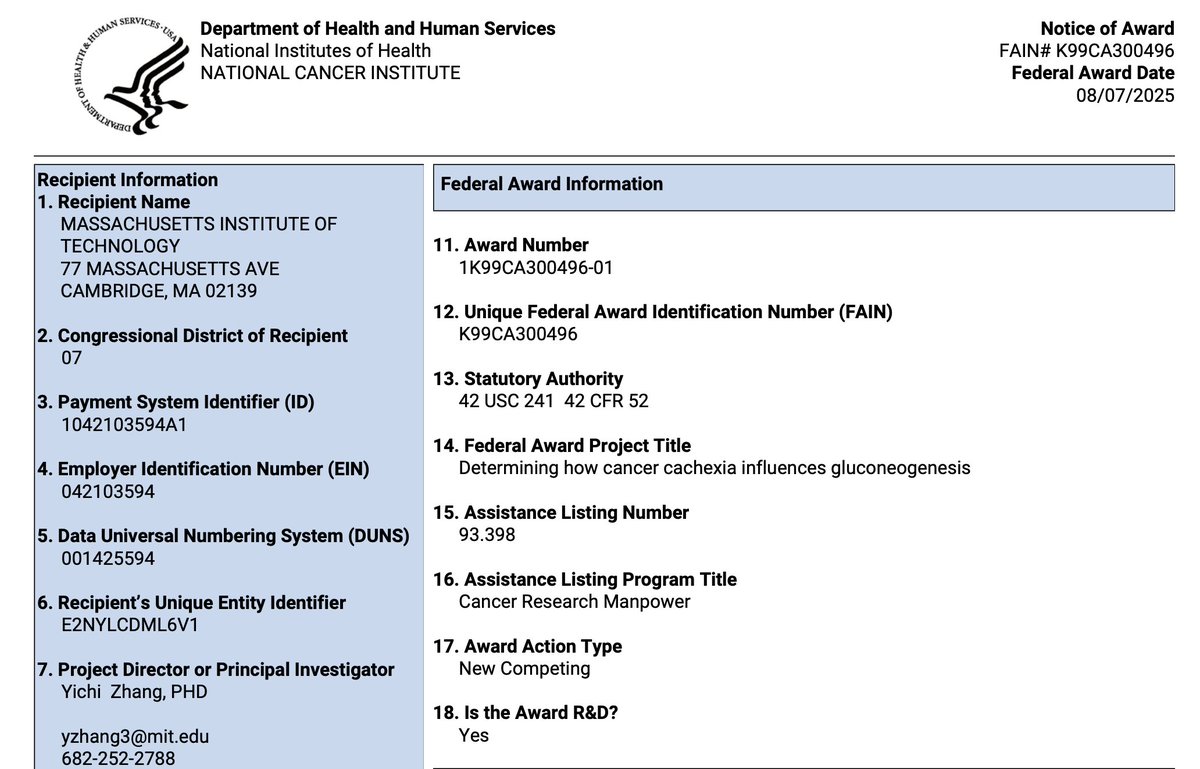
Happy to announce that I was awarded a pathway to independence award (K99) from the NCI/NIH today to work on studying whole-body metabolism driving cancer cachexia. None of this would be possible without support from Damon Runyon Cancer Research Foundation and mentorship from Eric N. Olson and mvhlab.