Aastha Pal
@apal9_pal
research @StanfordMed • @BU_tweets alum • prev @BMSnews 🧬💻💊
ID: 1484215668656558080
20-01-2022 17:26:19
84 Tweet
281 Followers
862 Following






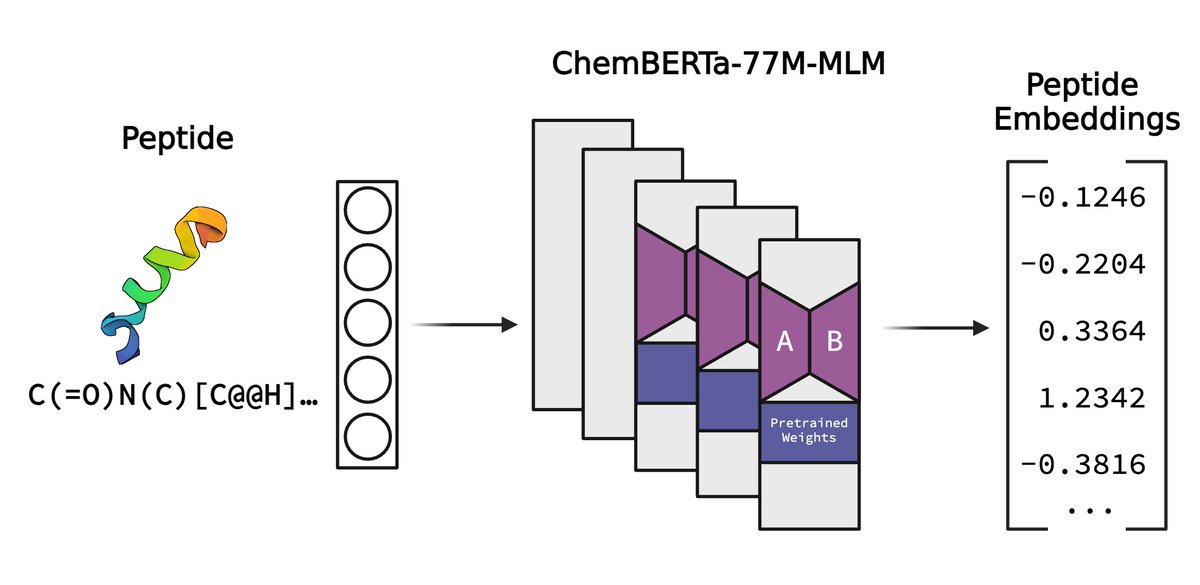
How can we better predict which T-cell receptors (TCRs) will bind to specific epitopes, a crucial challenge in understanding immune responses and developing immunotherapies? Stanford University "Sequence-based TCR-Peptide Representations Using Cross-Epitope Contrastive Fine-tuning of





How many proteins could be degraded by molecular glues? New paper from the team at Monte Rosa Therapeutics, Inc suggests it's more than previously thought! Study led by Georg Petzold Pablo Gainza-Cirauqui John Castle & others



After showing that AF2 can be used to design very large proteins by performing #RSO, I am happy to share another fun project we did: the #af2cycler Sergey Ovchinnikov @hendrik_dietz biorxiv.org/content/10.110…

During my sabbatical, one project I really enjoyed was training DNA foundation models with the team at Arc Institute. I've long believed that deep learning should unlock unprecedented improvements in medicine and healthcare — not just for humans but for animals too. I think this



Excited to share our joint work with Richard Shuai, Full-Atom MPNN (FAMPNN), a protein sequence design method that explicitly models both sequence and side-chain structure! 🧵 1/N




Excited too see nuclei.io featured on the cover of Nature Biomedical Engineering! AI-clinician collaboration paper rdcu.be/dLgV1