Dan Liu
@danliu_
PhD student in virology at MRC-University of Glasgow Centre for Virus Research @CVRinfo | Computational biology, virus-host interactions, LLMs 🦠 💻
ID: 867367172254736384
24-05-2017 13:10:23
22 Tweet
122 Followers
296 Following




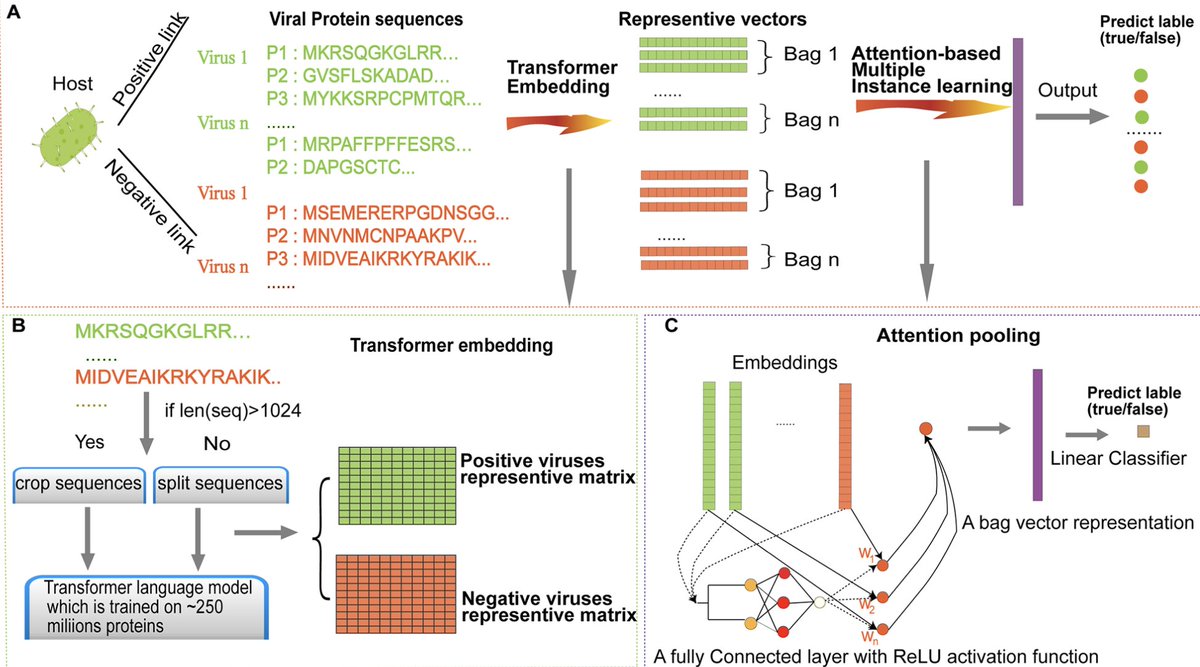
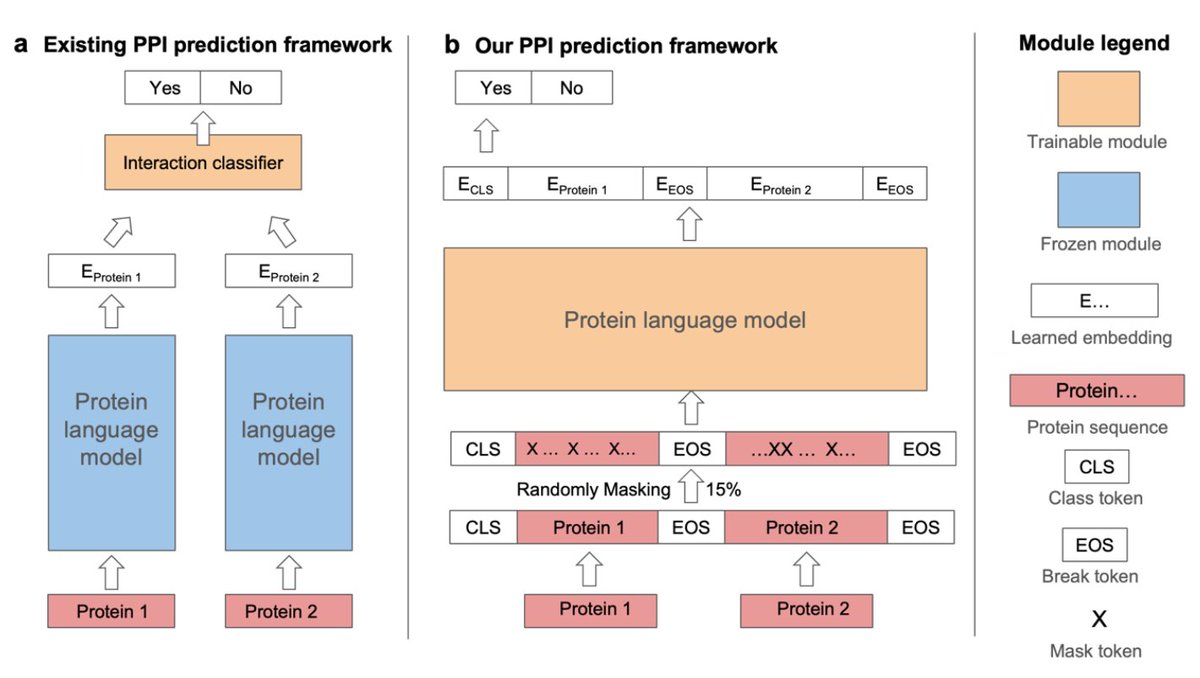
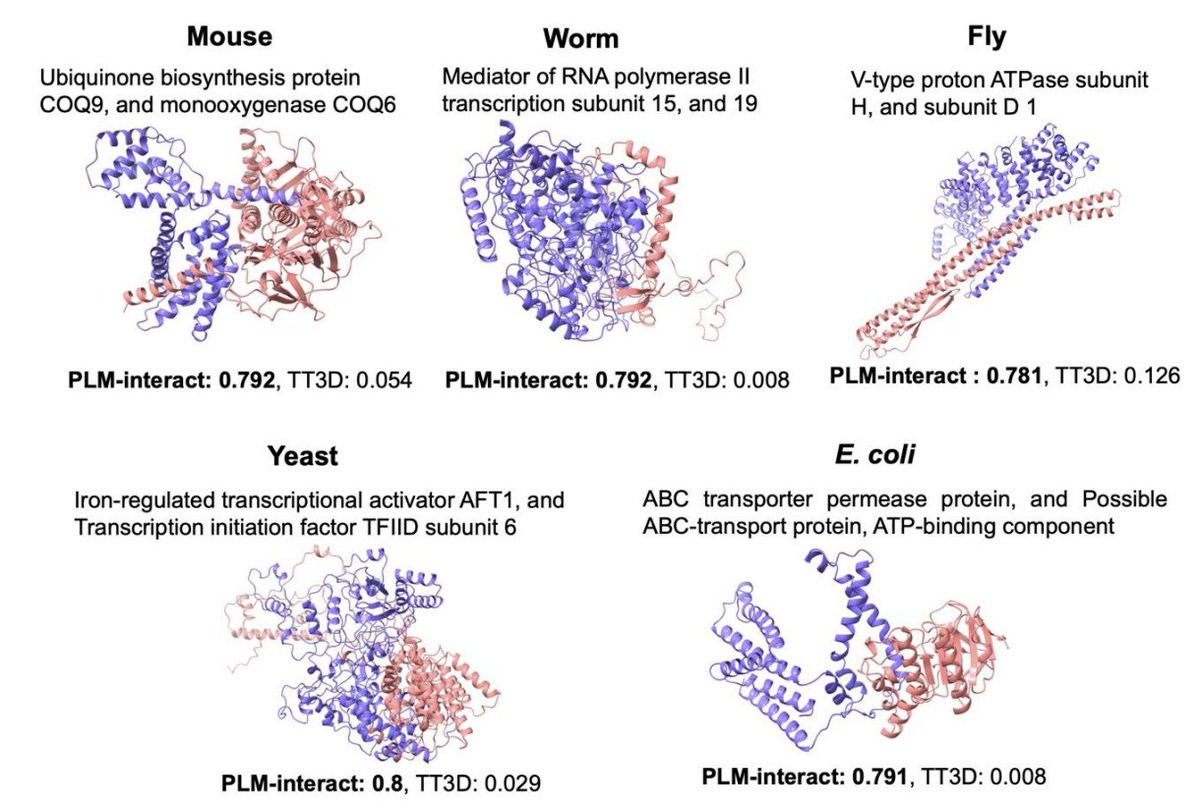
Prediction of virus-host associations using protein language models and multiple instance learning PLOS Comp Biol 1. EvoMIL introduces an innovative method for predicting virus-host associations by combining protein language models (PLMs) and attention-based multiple instance