Coby Viner
@cobyviner
Postdoc, Biology, UPenn
ID: 2796875636
https://www.cs.toronto.edu/~cviner/ 07-09-2014 23:58:16
1,1K Tweet
333 Takipçi
867 Takip Edilen






My posters are located next to each-other, with the (fewer) Q8/#KSChromatin23 posters, in the far back of Salon C. Looking forward to chatting about them! #KSChromatin23 #KSEpigenetics23 Keystone Symposia






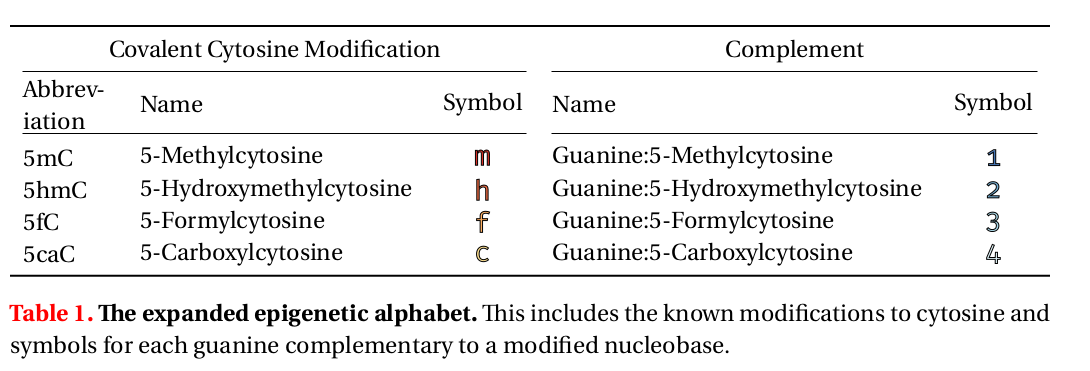
1/Now published Genome Biology! Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet. By Coby Viner Charles Ishak Nic Walker Hui Shi Marcela Sjöberg Shu Yi (Roxana) Shen David Adams Anne Ferguson-Smith Daniel De Carvalho Sarah Hainer et al! genomebiology.biomedcentral.com/articles/10.11…

Expanded DNA alphabet for gene regulation🧬 Beyond A/C/G/T, a study by Coby Viner and collaborators, led by Michael Hoffman @michaelhoffman.bsky.social University Health Network, added symbols like “m” and “h” to the genetic alphabet as a new way to study epigenetic regulation. >>genomebiology.biomedcentral.com/articles/10.11…


Very nice paper by @cobvinyer and colleagues in Michael Hoffman @michaelhoffman.bsky.social's lab. Methylation and Hydroxy-methylation decoding is really starting to be unpicked.