
Chirag Jain
@chirgjain
Assistant Professor @iiscbangalore. Previously @genome_gov, @GTcse and @IITDelhi
ID: 2784744199
https://at-cg.github.io 01-09-2014 21:15:23
177 Tweet
782 Followers
191 Following

RECOMB-Seq 2024 Call for Papers is out! recomb-seq.github.io/papers/ Submit via EasyChair. Select “RECOMBSeq 2024” track within RECOMB conference at easychair.org/my/conference?… Calendar: Abstract registration: January 30, 2024 Final submission: February 06, 2024 Event: April 27-28, 2024

Happy to see that the RAFT method was useful here Sudhanva Kamath github.com/at-cg/RAFT

The list of #RECOMB2024 Accepted Papers is now live: recomb.org/recomb2024/acc…. A huge congrats to all the authors! RECOMB Conference Series


Are nanopore UL reads only long reads we need? We developed Herro github.com/lbcb-sci/herro AI error correction model that can correct reads to accuracy above Q30 while trying to keep informative positions intact. w/ Dominik Stanojevic Dehui Lin Sergey Nurk 🇺🇦 Pore_XX_Singapore Oxford Nanopore


Join us at the second annual Symposium on Big Data Algorithms for Biology (BDBio) on May 31-June 1, 2024 IISc Bangalore The 2-day event will feature cutting-edge research in computational biology. Registration opens on March 20. More details here: bdbio.in


The CDS department at IISC Bangalore invites applications for its summer internship programme, aimed at students pursuing the relevant branch of engineering! The deadline for the application is 15th March 2024. cds.iisc.ac.in/call-for-cds-s… IISc Computational and Data Sciences #cdsinternship2024


Gapless assembly of complete human and plant chromosomes using only nanopore sequencing | bioRxivc Happy @keygene could contribute to this work. Oxford Nanopore #T2T #duolex biorxiv.org/content/10.110…


Our review paper on "Genome assembly in the telomere-to-telomere era" published online in Nature Reviews Genetics. It amazes me how much assembly has progressed in the past four years – often ~100X improvement in contiguity and base accuracy and with haplotypes and hard regions resolved!

We send our appreciation to the speakers, poster presenters, participants, student and staff volunteers who contributed to the resounding success of the BDBio 2024 held last week! IISc Bangalore IISc Computational and Data Sciences


Our recent work "Haplotype-aware sequence alignment to pangenome graphs" is now online at Genome Research (RECOMB'24 issue). It introduces new algorithms and complexity results for the problem. Led by Ghanshyam Chandra doi.org/10.1101/gr.279…




Happy to see our method for T2T genome assembly published! It addresses an important limitation of string graph, that is, the contained reads. Led by Sudhanva Kamath "Telomere-to-telomere assembly by preserving contained reads" doi.org/10.1101/gr.279…

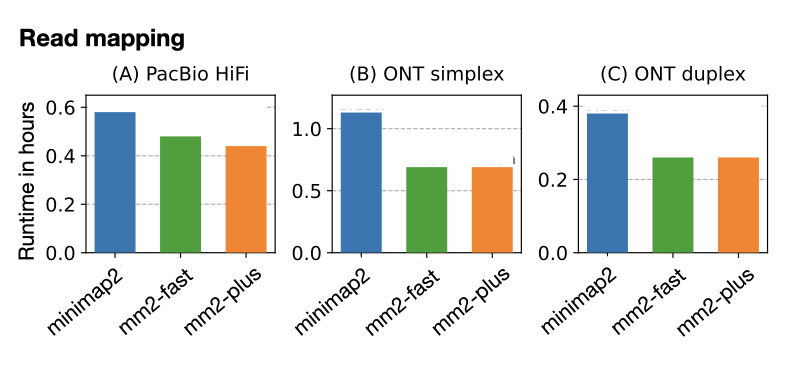
New method speeds up genome alignment of complete and near-complete genome assemblies in minimap2. Can also be useful for faster read mapping Github: github.com/at-cg/mm2-plus DOI: doi.org/10.1101/2024.1… Led by Ghanshyam Chandra, collab with Sanchit Misra (I am hiring) Mohd Vasimuddin Intel



The 3rd annual Symposium on Big Data Algorithms for Biology (BDBio) is happening this May! Join us for insightful talks and discussions on comp-bio topics Register, submit abstract, and see program details at bdbio.in IISc Bangalore IISc Computational and Data Sciences Centre for Brain Research, IISc


![IISc Computational and Data Sciences (@cdsiisc) on Twitter photo Symposium on Big Data Algorithms for Biology (May 31-June 1 2024) is now open for registration and abstract submission [Deadline: Apr 20] bdbio.in
Join us for the engaging talks, poster presentations, and panel discussions in various topics of comp. biology Symposium on Big Data Algorithms for Biology (May 31-June 1 2024) is now open for registration and abstract submission [Deadline: Apr 20] bdbio.in
Join us for the engaging talks, poster presentations, and panel discussions in various topics of comp. biology](https://pbs.twimg.com/media/GJVetHTWYAAkrHO.jpg)



