Rebekah Loving
@bound_to_love
I’m a weird, energetic, Hawai‘i girl
Mother & Wife
Computational biology, math, algorithms, software
PhD Candidate @ Caltech
ID: 1064373131500937216
19-11-2018 04:21:44
53 Tweet
172 Takipçi
118 Takip Edilen

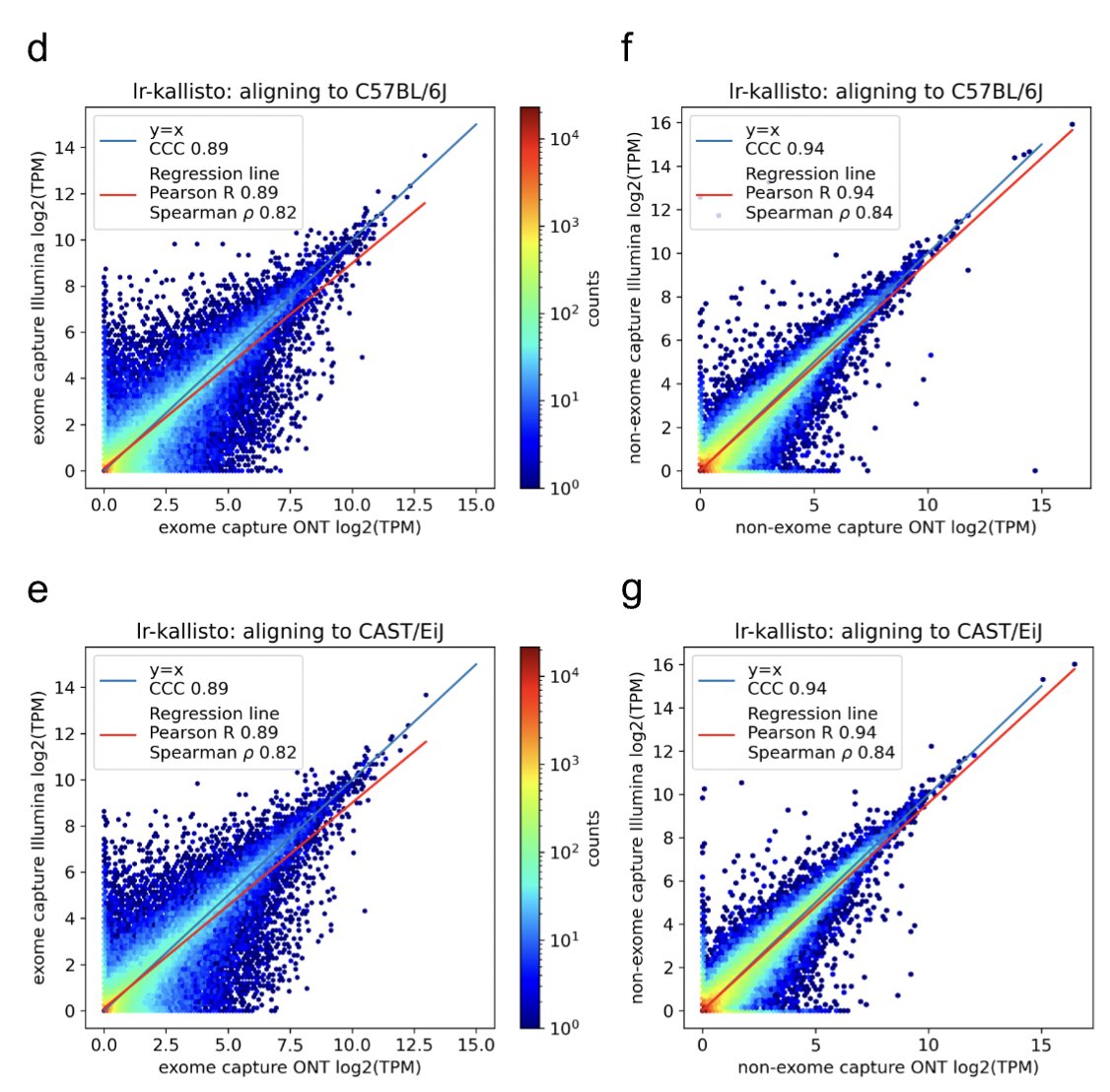
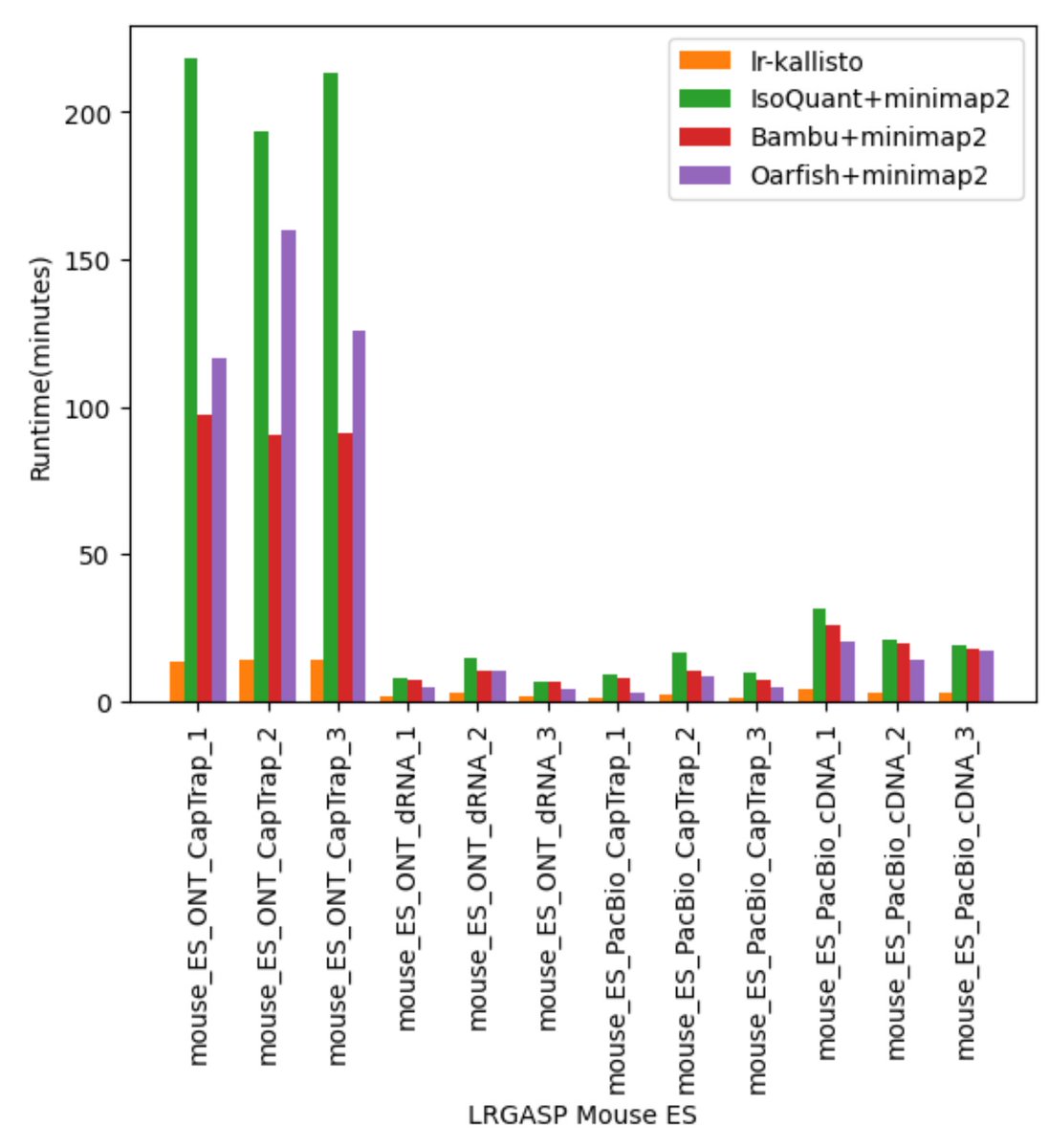
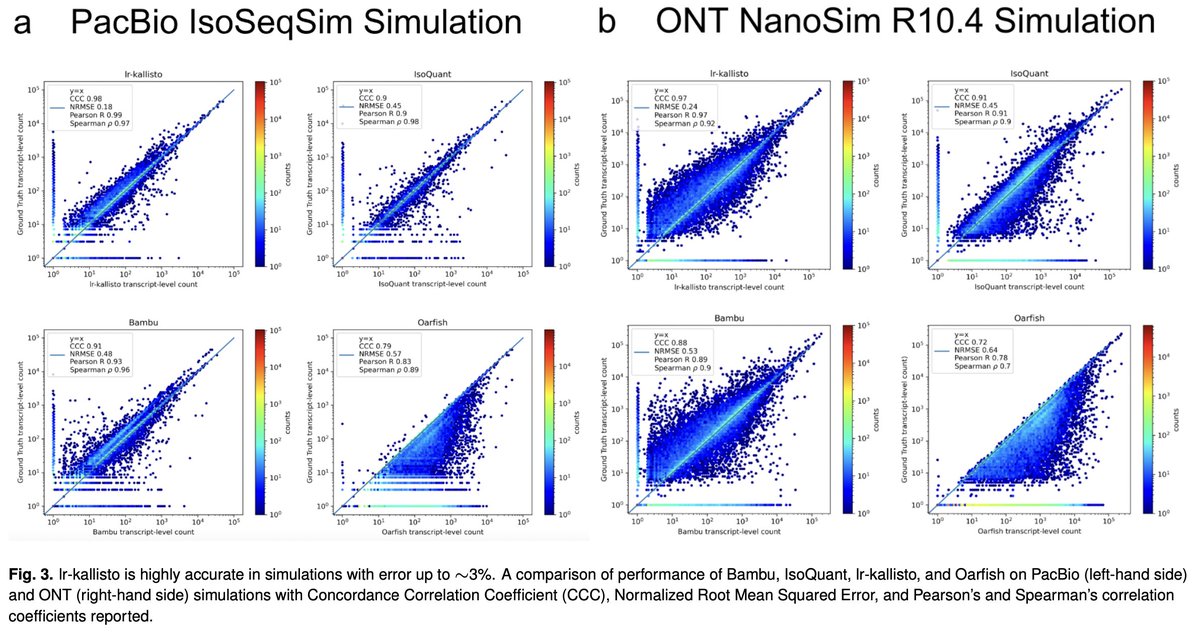
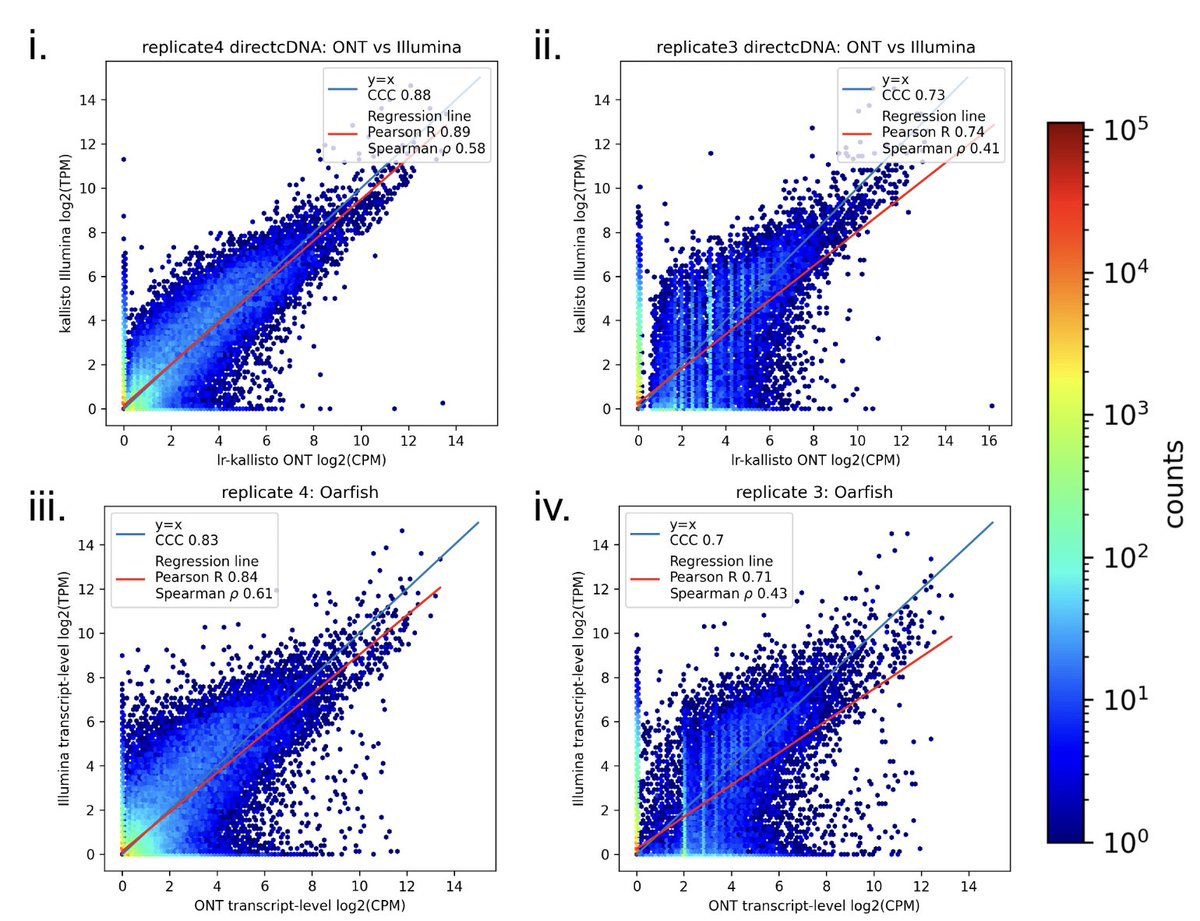
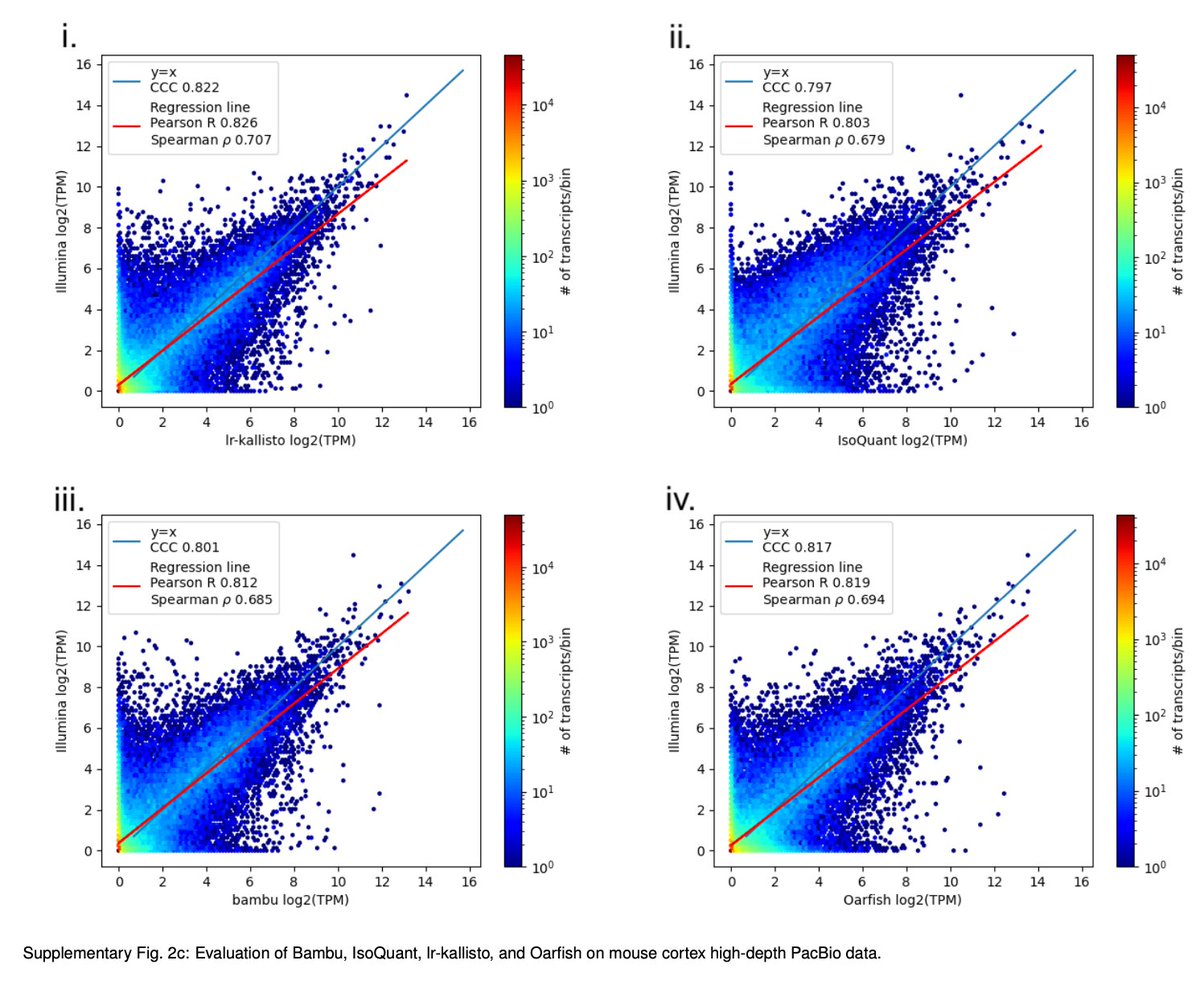
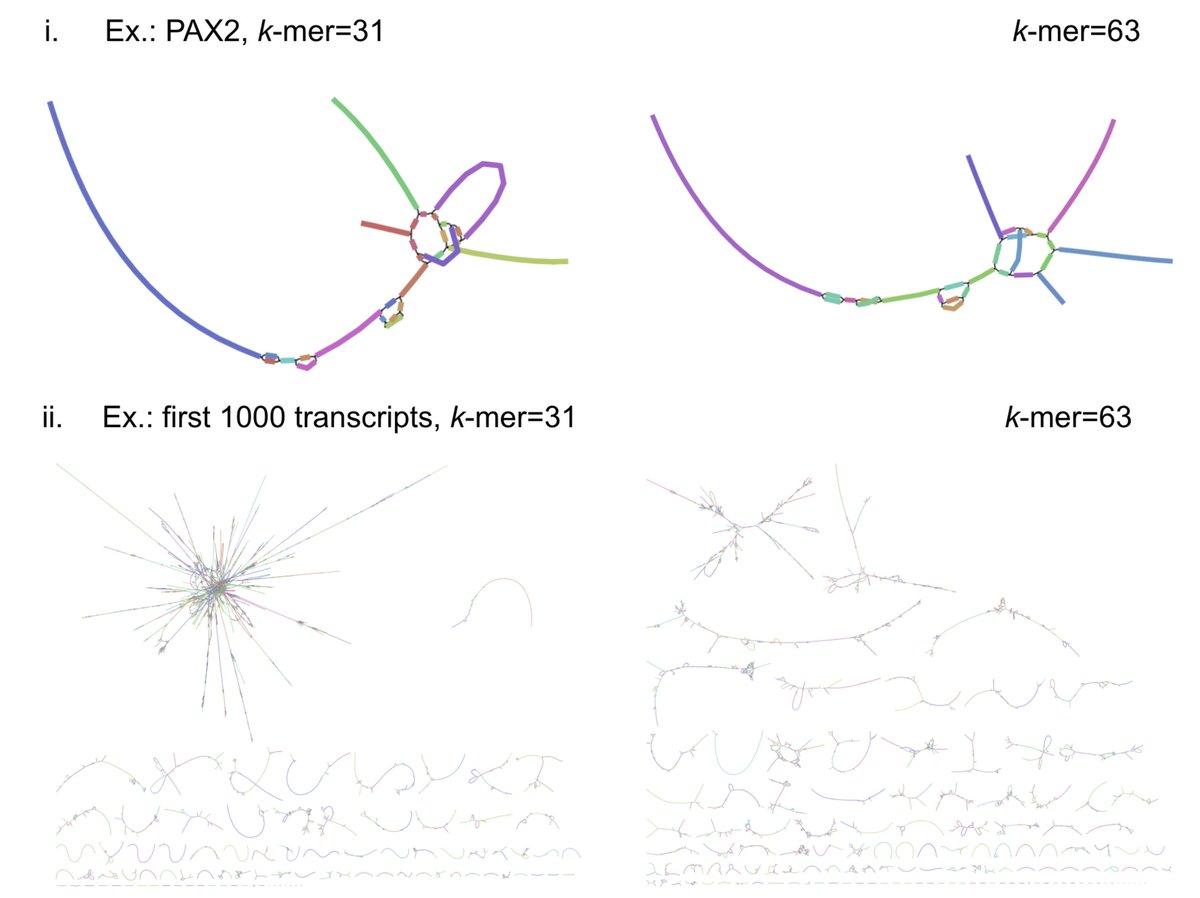
In a new work led by Rebekah Loving, joint with Ali Mortazavi and his lab, we introduce a method based on kallisto for accurate quantification of Oxford Nanopore or PacBio long-read RNA-seq, and demonstrate its accuracy with numerous comparisons and benchmarks 1/🧵 biorxiv.org/content/10.110…


We decided to adapt kallisto, which Delaney Sullivan has recently updated and improved in many ways, for long-read quantification. Rebekah Loving figured out that several modifications and improvement, especially w.r.t fragment lengths were needed. x.com/lpachter/statu… 3/

We needed a controlled dataset to benchmark on, so the Ali Mortazavi lab generated the ideal testbed: Illumina and Oxford Nanopore sequenced libraries with and without exome capture (to yield more reads from exons). And of course biological replicates for each. 4/
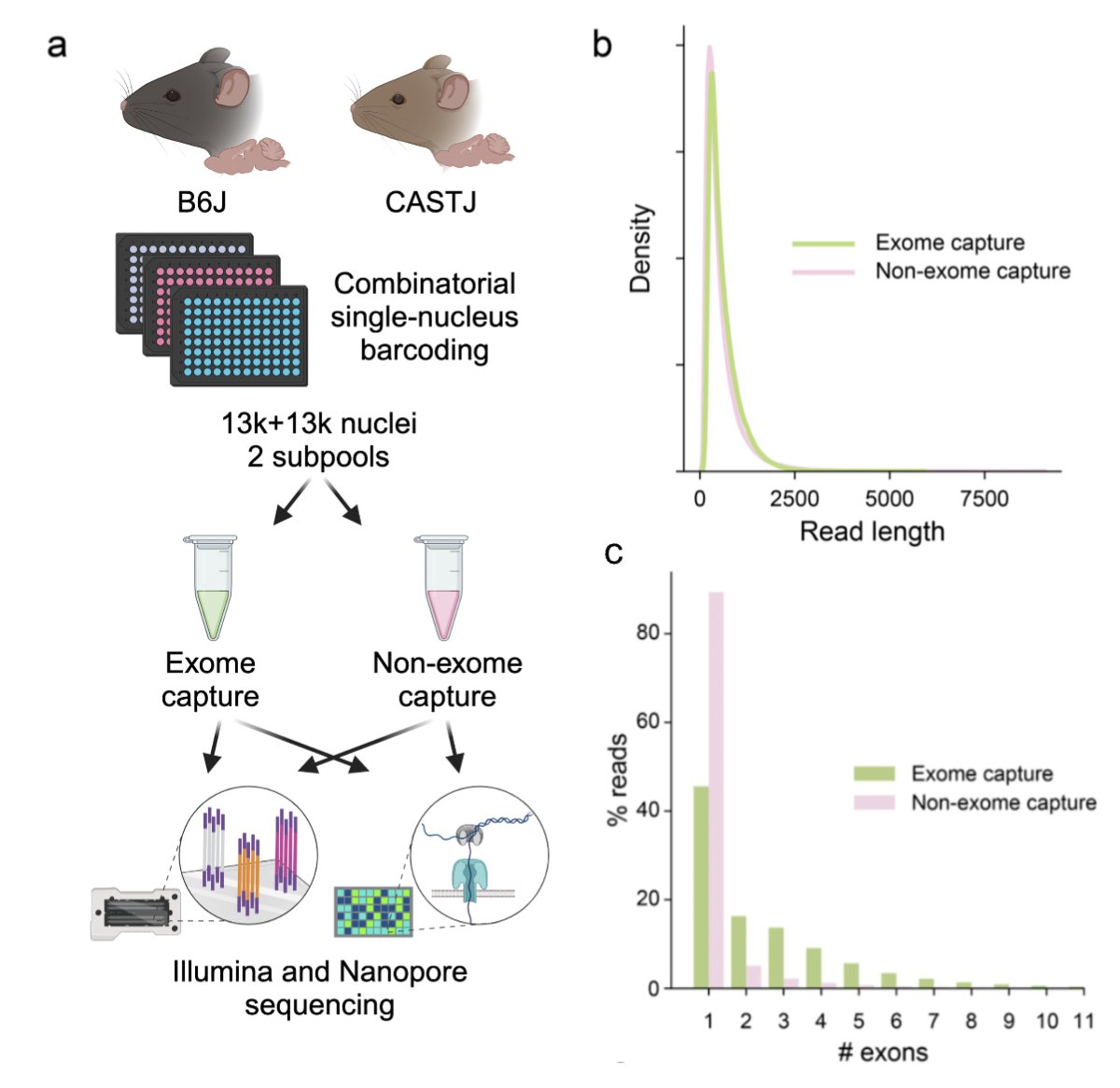








Many thanks to Barbara Wold, who also supervised the work, and the team from Ali Mortazavi lab team from UC Irvine who did exceptional work generating the matched ONT and Illumina data. An incredible accomplishment by Rebekah Loving! x.com/MarissaKawehi/… 12/



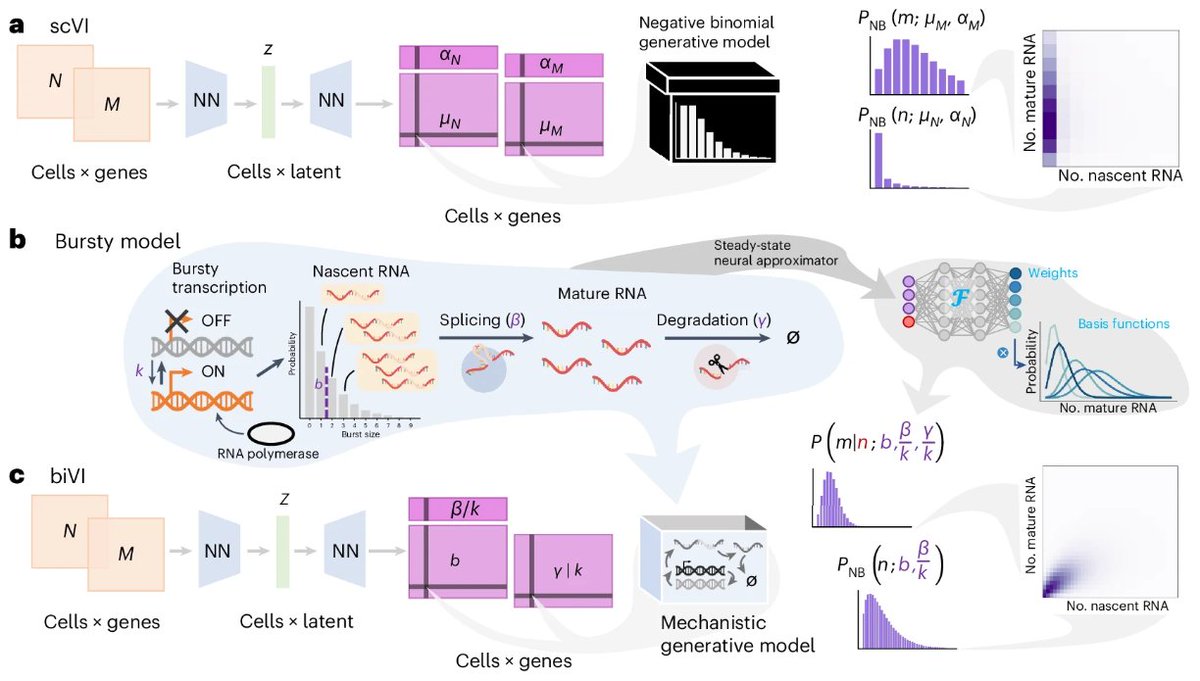
biVI models the biophysical processes generating nascent and mature single-cell transcriptomes using variational autoencoders. Maria Carilli Gennady Gorin Yongin Choi Lior Pachter nature.com/articles/s4159…

We are excited to welcome Dr. Lambda Moses, our new Postdoctoral Research Scientist! She specializes in spatial-omics data analysis and software development. In the bianca dumitrascu group, Lambda focuses on computational biology, single-cell genomics, and statistical bioinformatics.





Watch the talk from Joseph Rich, grad student in Lior Pachter's lab, from BioC2024 timestamp ~12:30 youtu.be/T6cOyKHlxpE?si…