Ana Sanchez-Fernandez
@ana_sanchezf
Machine learning for drug discovery, microscopy imaging data 🔬 | PhD student @jkulinz within the EU project @AiddOne
ID: 1560262451039727617
18-08-2022 13:49:09
28 Tweet
121 Takipçi
63 Takip Edilen




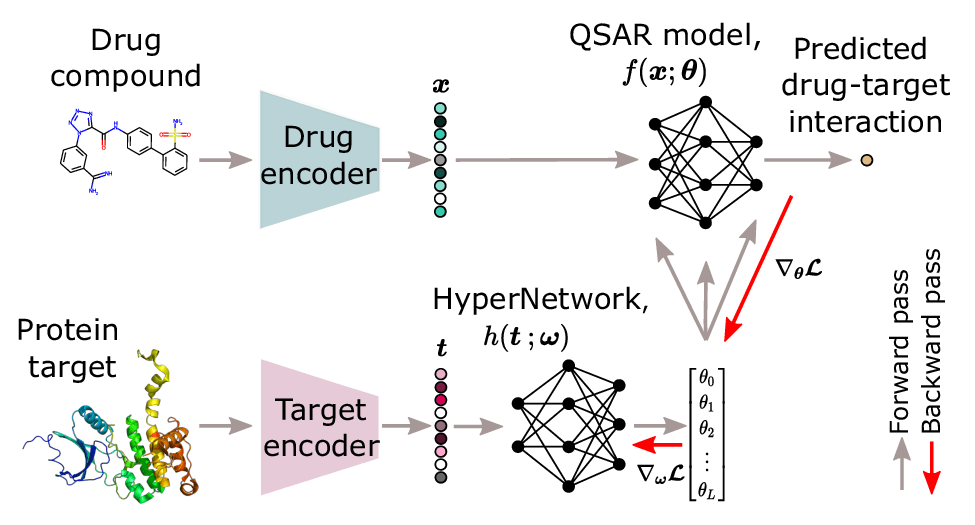
HyperPCM has been published in JCIM🧬 It's the first HyperNetwork dedicated to learning drug-target interactions with enhanced generalization to zero-shot tasks! 📰pubs.acs.org/doi/10.1021/ac… 🤗huggingface.co/spaces/emmas96… Work with Pieter-Jan Hoedt Sepp Hochreiter Günter Klambauer in https://bsky.app/profile/aidd.bsky.social.

VN-EGNN: E(3)-Equivariant GNNs with Virtual Nodes Enhance Protein Binding Site Identification New method to find binding pockets of proteins. Virtual nodes allow to employ distance losses directly. P: arxiv.org/abs/2404.07194 C: github.com/ml-jku/vnegnn 🤗:huggingface.co/spaces/ml-jku/…



ELLIS Machine Learning for Molecules workshop December 6, 2024, HYBRID Paper submission deadline: November 1st. Program chairs: Francesca Grisoni @jtmargraf an me Webpage: moleculediscovery.github.io/workshop2024/ We hope to see you all there!

Keynote at SETAC 2024 was a great opportunity to promote ai-dd.eu & https://bsky.app/profile/aichemist.bsky.social, e.g., contrastive learning by Ana Sanchez-Fernandez/Günter Klambauer, representation learning by Tox24 Challenge or #TransOrGAN published by our associated partner Weida Tong at SI of Chem. Res. Toxicol.