
Alexey Ruzov
@alexeyruzov
Epigenetics and epitranscriptomics
ID: 1211407375246098432
29-12-2019 22:03:39
1,1K Tweet
1,1K Followers
895 Following


I’m so thrilled to share my first first-author pre-print! 💪🎉🥳 This was an amazing collaborative work from Nitika Taneja, Andrés Aguilera and @lugerlab! How do #chromatinremodelers, #Rloops and mutations in #cancer correlate? A small 🧵below for you! biorxiv.org/content/10.110…


Curious about the seemingly contradictory functions of m6A in R-loops? Check out this comment by @a_abakir & Alexey Ruzov out in Nature Genetics where they propose a new model for a dual function of m6A in R-loop regulation. nature.com/articles/s4158…

Abdulkadir Abakir, PhD Nature Genetics Reik Lab Altos Labs Alexey Ruzov The dual function of m6A is really instersting!And not only existing in R-loop regulation, but also in gene expression regulation!


The SharedIt link for our (with Abdulkadir Abakir, PhD) comment in Nature Genetics rdcu.be/dUbLY We propose that in different contexts, m6A may either directly prevent R-loop accumulation or stabilize R-loops via the formation of RNA abasic sites. nature.com/articles/s4158…

Nice comment in Nature Genetics about the role of m6A in R-loop dynamics. Just one little detail: you refer to FLC (FLOWERING LOCUS C) as FLORAL REPRESSOR LOCUS. Plant people are always surprised that we´re still interested in it. There´s still work to do😜 nature.com/articles/s4158…


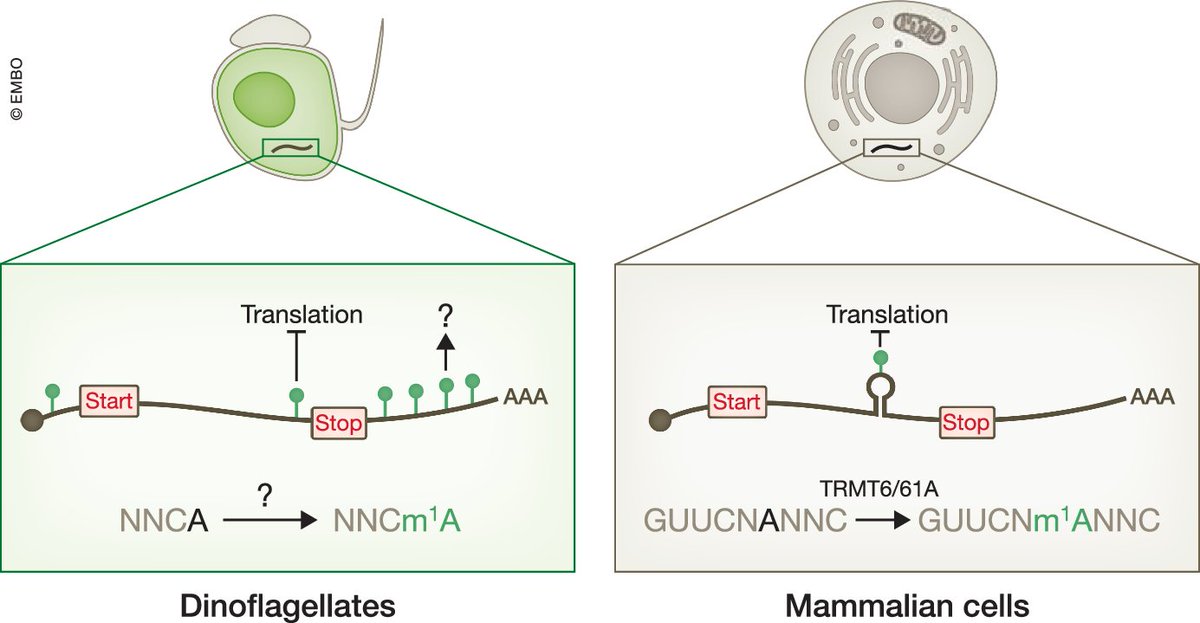
N1-methyladenosine (#m1A) is abundant and dynamic in the mRNAs of #dinoflagellates, and could have an important role in post-transcriptional #generegulation. Paper by Hao Chen and co-workers: embopress.org/doi/full/10.10… and the highlight by JaffreyLab: embopress.org/doi/full/10.10…





Now in print! A model for a dual function of N6-methyladenosine in R-loop regulation | Nature Genetics Nature Genetics nature.com/articles/s4158…

exciting future for m6A in #Rloop great perspective Abdulkadir & Alexey Ruzov so much yet to learn 🤓

Excited to share our latest research out now in Molecular Cell. DNA supercoiling creates topological issues that affect nuclear DNA transactions, with cells using topoisomerases to resolve these constraints. (1/7) cell.com/molecular-cell…



Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment. nature.com/articles/s4158…

Pleased to share our latest work: Chimeric cofactors enable methyltransferase-catalyzed prenylation: Chem cell.com/chem/fulltext/… Kudos to @EnzymeNicoCorn, Arne Hoffmann, Pulak Ghosh and x-less coathors 👏🤩👍

