Shihao
@shihaofeng18
ID: 1269560569478037509
07-06-2020 09:23:34
28 Tweet
26 Takipçi
130 Takip Edilen





Roughly one year ago we started out venturing in the world of #proteindesign. Today I am very happy to share our first preprint ‘Efficient and scaleable de-novo protein design using a relaxed sequence space’ (biorxiv.org/content/10.110…) with Sergey Ovchinnikov & @hendrik_dietz 1/x

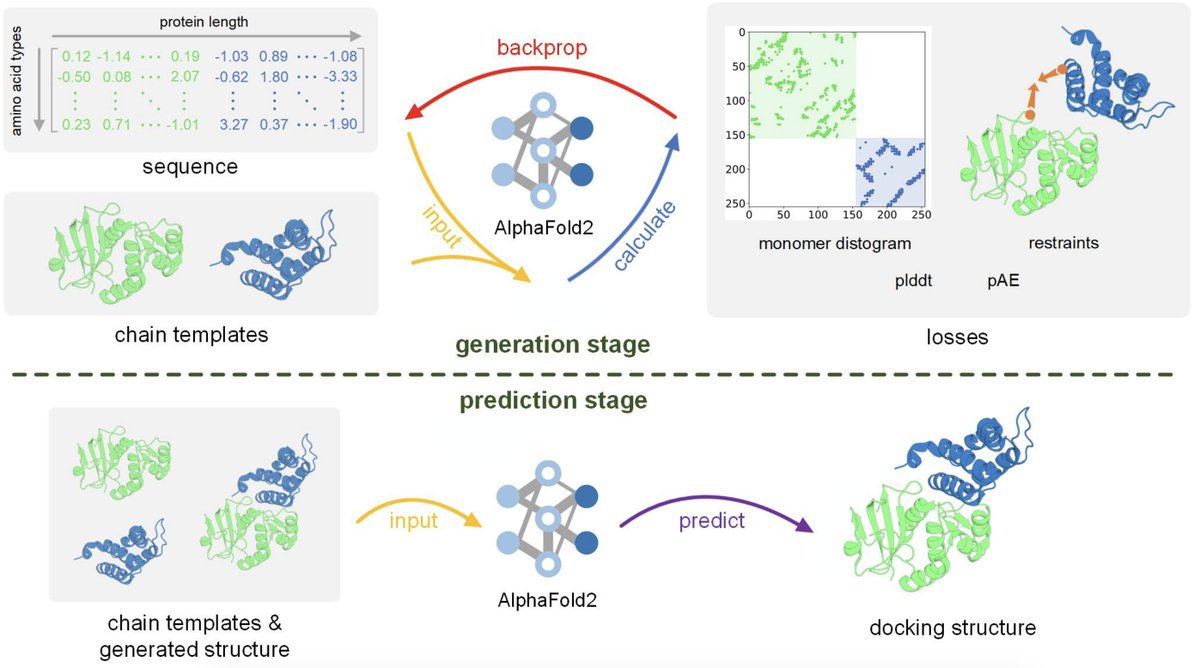
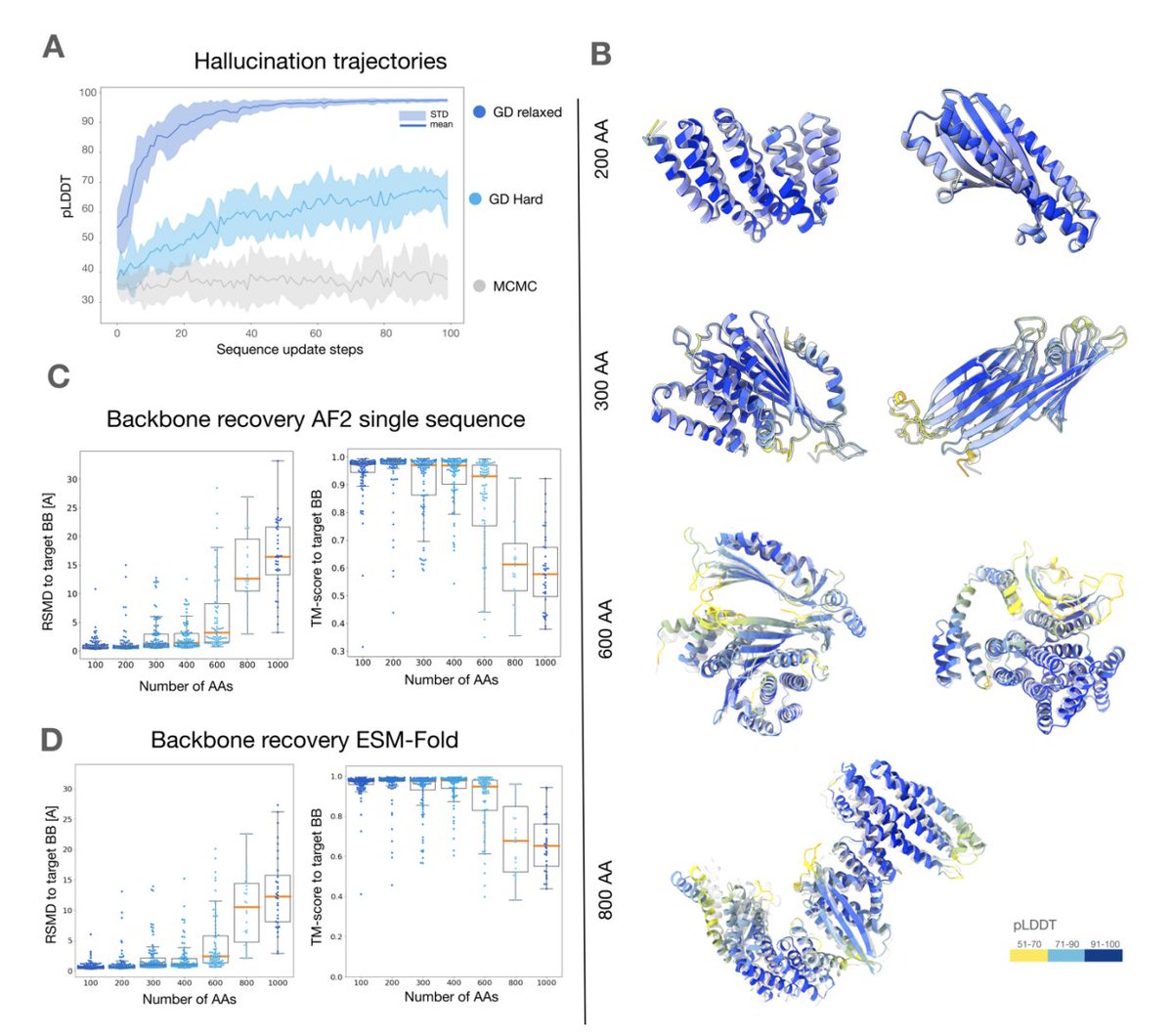
AlphaFold inverted to hallucinate denovo proteins of up to 600 amino acids in length🤯 (animation below shows the designed protein docked into CryoEM density) Exciting work with: Christopher Frank, Ali Khoshouei, Yosta de Stigter, Dominik Schiewitz, Shihao, @hendrik_dietz


Our new work with Sergey Ovchinnikov Institute for Protein Design core labs, DiMaio lab. We introduce AfCycDesign, where we adapt the #AlphaFold network for cyclic peptide structure prediction and design. biorxiv.org/content/10.110…

There are other protein designers, but I think Sergey Ovchinnikov is the only protein magician. This is the latest in a series of genius hacks (and I mean this in the best possible spirit of the term) from him and his collaborators, in this case Gaurav Bhardwaj & team at Institute for Protein Design.

Efficiently generate de novo proteins by - optimizing residue logits for max AF confidence - redesigning the sequence using ProteinMPNN Tested in the lab, including CryoEM structures Christopher Frank Ali Khoshouei Sergey Ovchinnikov @hendrik_dietz biorxiv.org/content/10.110…




I'm excited to share that I'll be joining MIT Biology as an Asst Prof. in Jan 2024! Come join us! 🤓🧪🖥️🧬


Now out in slightly shorter form :) nature.com/articles/s4158… Kotaro Tsuboyama Justas Dauparas Elodie Laine Sergey Ovchinnikov



![Amirhossein Kazemnejad (@a_kazemnejad) on Twitter photo 🚨Stop using positional encoding (PE) in Transformer decoders (e.g. GPTs). Our work shows 𝗡𝗼𝗣𝗘 (no positional encoding) outperforms all variants like absolute, relative, ALiBi, Rotary. A decoder can learn PE in its representation (see proof). Time for 𝗡𝗼𝗣𝗘 𝗟𝗟𝗠𝘀🧵[1/n] 🚨Stop using positional encoding (PE) in Transformer decoders (e.g. GPTs). Our work shows 𝗡𝗼𝗣𝗘 (no positional encoding) outperforms all variants like absolute, relative, ALiBi, Rotary. A decoder can learn PE in its representation (see proof). Time for 𝗡𝗼𝗣𝗘 𝗟𝗟𝗠𝘀🧵[1/n]](https://pbs.twimg.com/media/FxiwwRtaQAkKi_8.png)