Maria Puschhof
@mariacheinz
Computational Scientist at HI-STEM / DKFZ
Heterogeneity in cancer | Computational Biomedicine
ID: 1482828753180975105
16-01-2022 21:35:08
87 Tweet
186 Takipçi
284 Takip Edilen


Wonder how much colorectal cancer organoids could be improved by reconstructing the tumor microenvironment? Check out our latest pre-print and opus magnum by Nicolas Broguiere and colleagues!



Extremely honoured to receive the ERC Consolidator this year #ERCCoG. Looking forward to the next 5 years of exciting science to unravel the intricacies how tumors become malignant. #CancerPrevention @UMCU_CMM Oncode Institute

Very happy to announce that our research on the microbiome in T -cell immunotherapies will receive this incredible boost . Thanks to the lab TMS lab, my mentors Eran Elinav, Marcel van den Brink, Roland Schmid, Nisar Malek, and institutions @M3 DKFZ NCT Heidelberg Gastroenterology@TUM Controlling Microbes to Fight Infections

🧬Explore disease at a multicellular level, capturing variations in patient samples across cell types, tissue features, studies, and technologies with Multicellular Factor Analysis, peer-reviewed by Review Commons and now published eLife - the journal elifesciences.org/articles/93161




Curious about inferring cell-cell communication in space or in your multi-modal dataset? Check out LIANA+, the latest framework from the Saez-Rodriguez Group. Congrats to @DanielBDimitrov and all the co-authors!

Still time until 15th September to submit an abstract to join us in Berlin in the European Society for Spatial Biology conference 👇


Fantastic news for the #microbiome-related #cancer field! Exciting research is continuing at Epithelium Microenvironment Interaction Lab @ DKFZ, now supported by #ERCStG. Many congrats to Jens Puschhof and the entire team! Can’t wait to this awesome team revolutionizing how we think about #bacteria in #cancer...


Looking for a job as a #Research #Technician in #Cancer #Microbiome? We offer a collaborative team Epithelium Microenvironment Interaction Lab @ DKFZ , diversity of methods, opportunity for further training and a fun work place at DKFZ! Apply now (or share with your network): jobs.dkfz.de/en/jobs/167181…

Hyped to be at the #ESSB European Society for Spatial Biology #spatial #biology conference together with some of my colleagues from the Saez-Rodriguez Group . Great science, beautiful venue, looking forward to the next 1.5 days...

Looking forward to connect with the local single cell enthusiasts in Heidelberg tomorrow! Thanks a lot for the invitation Saez-Rodriguez Group Julio Saez-Rodriguez Natalia Gabrielli and maria!


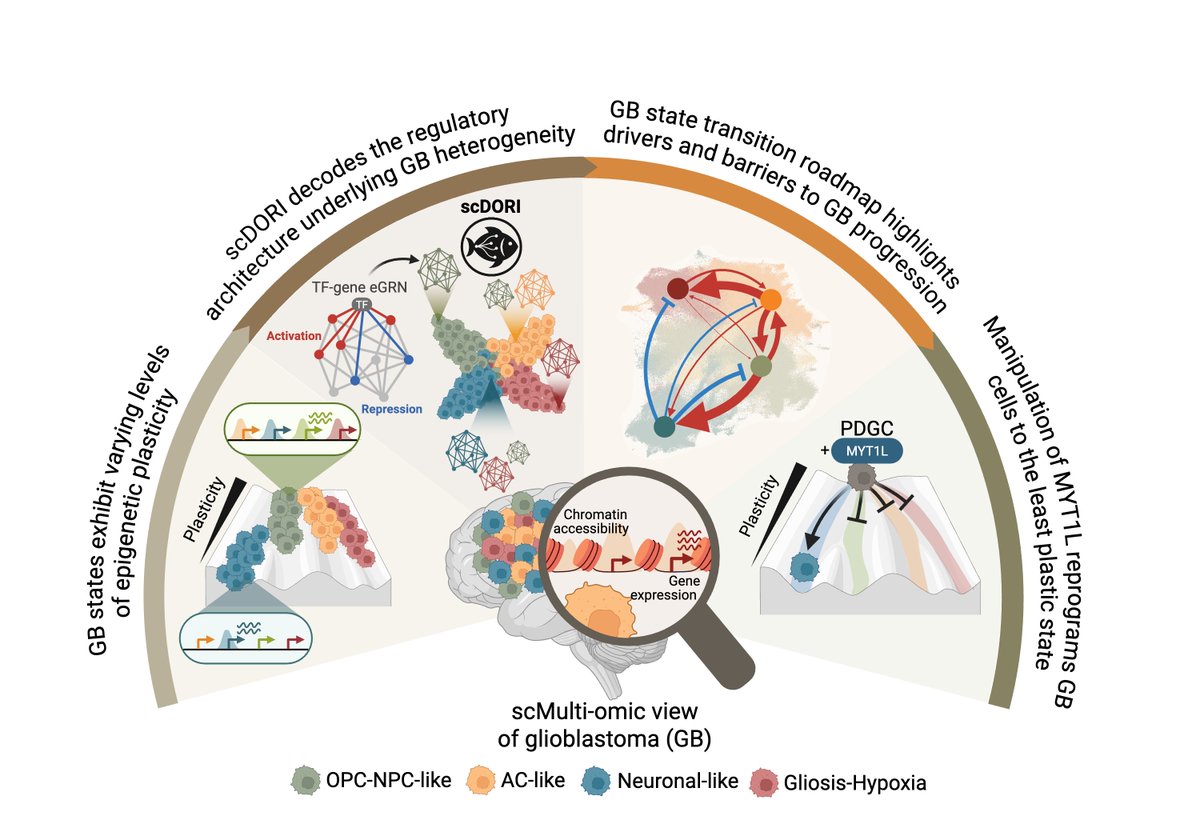
🧠 Excited to share my main PhD project! We mapped the regulatory rules governing Glioblastoma plasticity using single-cell multi-omics and deep learning. This work is part of a two-paper series with Omer Ali Bayraktar , Oliver Stegle and Moritz Mall groups. Preprint at end🧵👇