
Cistrome Data Browser
@cistromedb
A portal for browsing public #ChIPseq, #DNaseseq and #ATACseq datasets with analysis and visualization functions. #Epigenetic #GeneRegulation
ID: 1216563164814307328
http://cistrome.org/db/ 13-01-2020 03:30:45
40 Tweet
215 Followers
77 Following

Very excited to share SELMA, a computational method we developed for ATAC-seq & DNase-seq bias estimation, recently published in Nature Communications spearheaded by Shengen Shawn Hu w input from Cliff Meyer Lin Liu @guertinlab Ke Deng & Tingting Zhang nature.com/articles/s4146…



Our work on Cistrome Explorer has been published! It lets you analyze thousands of chromatin profiles together with cell types, hierarchical clustering, and any other metadata, as well as information queried from Cistrome Data Browser doi.org/10.1093/bioinf… cisvis.gehlenborglab.org



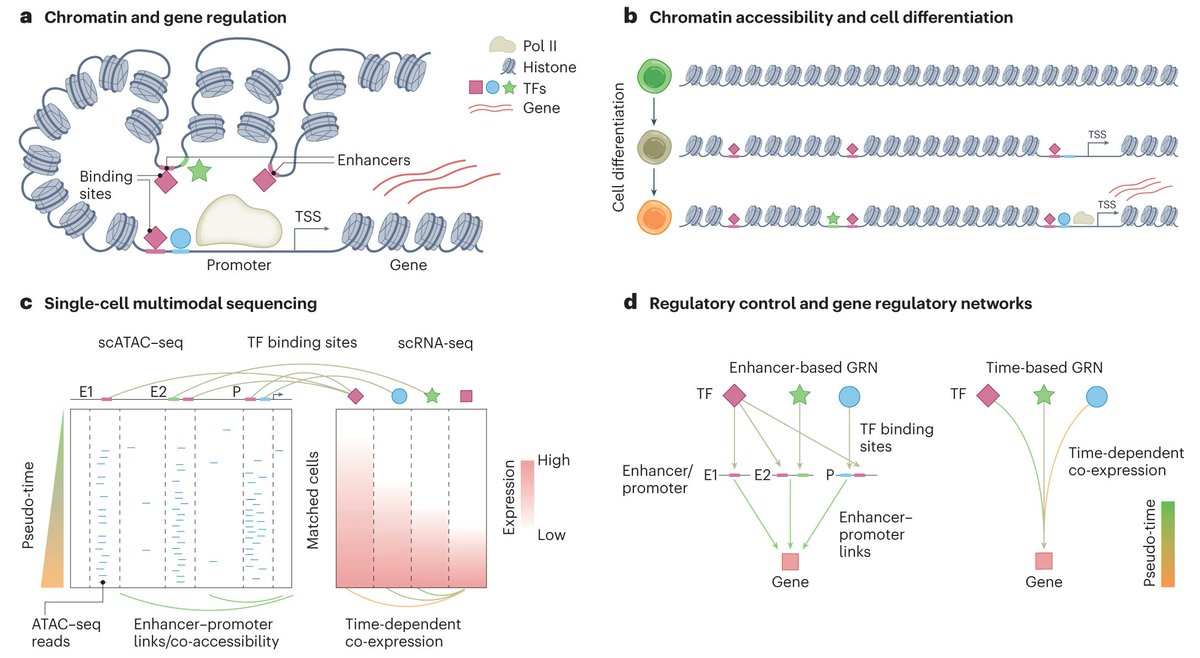
A short take on how computational approaches leverage multimodal single cell (RNA+ATAC) for inference of regulatory networks and a brief review of recent ground-breaking work by Dictys, SCENIC+ and MIRA by Luca Pinello @steinaerts Cistrome Data Browser et al. rdcu.be/disS3

.Cistrome Data Browser Dana-Farber Data Science disentangle technical and biological effects, facilitating batch-confounded chromatin and gene expression state discovery & enhancing the analysis of perturbation effects on cell populations. #BiotechNatureComms nature.com/articles/s4146…