Zichen Wang
@zichenwangphd
ML Scientist @awscloud. #MachineLearning, Healthcare, Biology. Formerly PhD & research faculty @IcahnMountSinai. Views are my own.
ID: 713877589
https://wangz10.github.io 24-07-2012 07:33:23
249 Tweet
261 Takipçi
445 Takip Edilen


Check out the great work from our amazing intern Yijun Tian








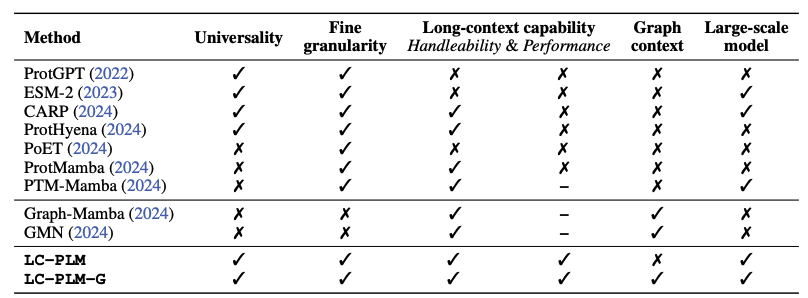
A long-context protein language model with a bidirectional Mamba architecture. Yingheng Wang Zichen Wang, Gil Sadeh, Luca Zancato, Alessandro Achille, George Karypis Huzefa Rangwala